August 2019
Clostridium difficile (C. diff) is an opportunistic pathogen that establishes in the colon when the gut microbiota are disrupted, often seen in seriously ill or elderly patients. Clostridium difficile infection (CDI) has become the most common cause of antibiotic-associated diarrhea and gastroenteritis-associated death in developed countries, accounting for a half-million cases and 29,000 deaths annually in the US. It is considered an “urgent threat” by the CDC.
The pathology of CDI is primarily mediated by homologous exotoxins, TcdA and TcdB. While the relative roles of these two toxins in the pathogenesis of CDI are not completely understood, recent studies have shown that TcdB is more virulent than TcdA and more important for inducing the host inflammatory and innate immune responses.
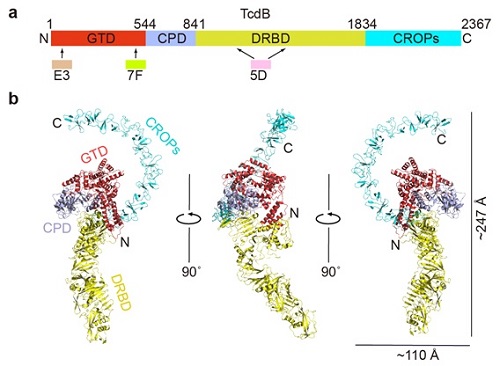
Figure. 1. Overall structure of the full length TcdB holotoxin. (a) A schematic diagram showing the domain organization of TcdB and the approximate VHH-binding regions. GTD, glucosyltransferase domain (red); CPD, cysteine protease domain (light blue); DRBD, delivery and receptor-binding domain (yellow); CROPs, combined repetitive oligopeptides domain (blue). (b) Cartoon representations of TcdB holotoxin. The 3 VHHs that were used to facilitate crystallization were omitted for clarity. The TcdB domains are colored the same as that shown in panel (a).
Both TcdB and TcdA are large molecular weight proteins with multiple domains and complicated conformational flexibility. Numerous structures have been reported for fragments of TcdA and TcdB, which have provided tremendous insight into the functions of these toxin domains. However, it remains unknown how individual domains interact within the supertertiary structure of the holotoxin and how the holotoxin dynamically responds in a precise stepwise manner to the environmental and cellular cues that lead to intoxication.
A recent study, led by researchers from the University of California, Irvine (UCI), uncovered the long-sought-after, three-dimensional structure of a toxin primarily responsible for devastating Clostridium difficile infection (Fig. 1). Published in Nature Structural & Molecular Biology, the study sheds light on the weaknesses of TcdB, one of the toxins secreted by the Clostridium difficile bacteria and the main cause of CDI.
The team determined the crystal structure of the toxin by collecting macromolecular crystallography data at the Northeastern Collaborative Access Team 24-ID-C beam line at the Advanced Photon Source (APS) and at Beam Line 9-2 at the Stanford Synchrotron Radiation Lightsource (SSRL). To further probe the structural dynamics of the toxin, the team used a combination of small-angle x-ray scattering (SAXS), single-molecule fluorescence resonance energy transfer (smFRET), and cross-linking mass spectrometry (XL-MS), of which the small-angle x-ray scattering experiments were performed at SSRL Beam Line 4-2.
This study revealed the 3D structure of the gigantic TcdB holotoxin at a near atomic resolution for the first time and indicated the pH-dependent structural flexibility of TcdB that may help to modulate its activity in response to environmental pH change. through this studty the team also demonstrated how three antibodies could neutralize TcdB, revealing intrinsic vulnerabilities of the TcdB toxin that could be exploited to develop new therapeutics and vaccines for the treatment of CDI.
Standard treatment for CDI involves using broad spectrum antibiotics that reduce the level of C. diff bacteria. However, the treatment also kills the good bacteria in the gut and disrupts the normal gut microbiome. This approach often leads to frequent disease recurrence.
More potent and cost-effective therapies for CDI remain a desperate need. The 3D structure of TcdB literally provides a blueprint for the development of next-generation vaccines and therapeutics that have enhanced potency and broad-reactivity across different C. diff strains. The UCI team has already begun working on a novel vaccine based on the new structure with early studies showing promising results.
