May 2013
Aminoacyl-tRNA synthetases are required in all three domains of life to add the correct amino acid to its cognate tRNA, an essential step in the process of protein synthesis. In eukaryotes and some bacteria, the traditional pathway of aminoacylation exists for glutamine, in which glutaminyl-tRNA synthetase (GlnRS) binds to tRNAgln, glutamine and ATP and first forms a glutaminyl adenylate molecule that is then covalently attached to the 3’-end of tRNAgln with the release of AMP. In most bacteria and all archaea, a different pathway exists where a non-discriminating glutamyl-tRNA synthetase (GluRS) attaches glutamic acid to both tRNAglu and tRNAgln. The misacylated glu-tRNAgln is then converted to gln-tRNAgln by the GatCAB amidotransferase enzyme in bacteria and some archaea, or by the GatDE amidotransferases in other archaea.
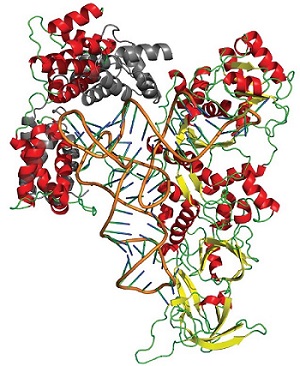
The eukaryote Saccharomyces cerevisiae contains a specific glutaminyl-tRNA synthetase (ScGlnRS), which is an 809-residue protein that contains the signature motifs found in all Class I tRNA synthetases as well as a 215-residue domain appended to its N-terminus. This domain is nearly ubiquitous among eukaryotic GlnRS species, but is absent in prokaryotic homologs; indeed, eukaryotic tRNA synthetases have often been shown to contain additional domains appended to their N-terminal or C-terminal ends, compared to their prokaryotic homologs. Some of these domains are known to be involved in various roles, including nucleic acid binding, protein-protein interactions, and hydrolytic editing mechanisms, but the functions of many remain uncertain.
Using remotely collected data from SSRL a research group led by Edward Snell of the Hauptman-Woodward Medical Research Institute previously described the structure of the N-terminal domain (NTD) of ScGlnRS, revealing that it has an extraordinary structural resemblance to the region of the B subunit of the GatCAB amidotransferase that binds to tRNAgln. Although structural data for two prokaryotic GlnRS species exists, no structure has been reported for any full-length eukaryotic GlnRS. Using SSRL macromolecular crystallography Beam Line 11-1 Snell's group also determined the structure of the C-terminal domain (CTD) of ScGlnRS from crystals of full-length GlnRS. Based on this structure, on the structure of the NTD, and on small angle x-ray scattering (SAXS) data of the full-length enzyme measured at SSRL Beam Line 4-2, they have developed a model of the full-length enzyme in solution. Using crystallographic structures and homology with known transamidosome and GlnRS-tRNA complex structures, they also modeled the full-length enzyme bound to tRNAgln . The combined results suggest that C-terminal domain binding to tRNA results in a large conformational reorientation of the N-terminal domain allowing for interactions between the N-terminal domain and the tRNA. The N-terminal domain plays a direct role in tRNA binding.
The combination of crystallographic and solution SAXS studies enabled by SSRL facilities has yielded fundamental new insights into the structural rearrangements occurring in eukaryotic GlnRS-tRNAgln complex formation.
