The omicron variant of COVID-19 was identified in the fall of 2021. It stood out from all of the other variants because of the many mutations that simultaneously occurred in its spike protein1. So far, surveillance and bioinformatics have been the main scientific tools in tracking COVID-19 evolution. Eventually, however, understanding COVID-19 evolution comes down to understanding the functions of key viral mutations. This is where structural biology kicks in and plays a critical role in tracking COVID-19 evolution.
In a study recently published in the journal Proceedings of National Academy of Sciences USA, Dr. Fang Li and colleagues at the University of Minnesota determined the high-resolution crystal structure of the omicron strain’s spike protein and its mouse receptor (Fig. 1A)2, using macromolecular cystallography x-ray data measured at Beam Line 12-1 of SSRL. Through detailed analysis, the researchers identified three mutations (Q493R, Q498R, and Y505H) in the omicron spike protein that are specifically adapted to two residues (Asn31 and His353) in the mouse receptor (Fig. 1B, 1C). After searching all of the available receptor sequences in the database, the researchers found that only the receptor from mice contains Asn31 and His353, while the receptors from several other rodent species contain one but not both Asn31 and His353. Thus, the researchers hypothesized that rodents, particularly mice, played a role in the omicron evolution. In contrast, these three mutations in omicron are structurally incompatible with the corresponding two residues (Lys31 and Lys353) in the human receptor (Fig.1D, 1E)2, further suggesting that non-human animal reservoirs facilitated the omicron evolution.
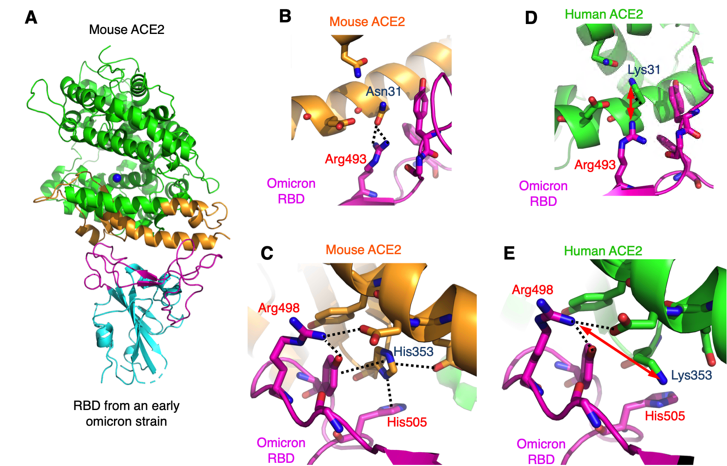
Based on the above structural findings, the researchers discussed a possible scenario in which humans might have transmitted the COVID-19 virus to non-human animals, which became new reservoirs for the virus and later transmitted the virus back to humans with novel mutations. The study suggests that surveillance of COVID-19 in rodents near human habitats and animal farms may help prevent new COVID-19 variants from emerging.
- S. K. Saxena et al., “Characterization of the Novel SARS-CoV-2 Omicron (B.1.1.529) Variant of Concern and Its Global Perspective”, J. Med. Virol. 984, 1738 (2022) doi:10.1002/jmv.27524.
- Q. Geng et al., “Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1”, J. Virol. 96, e0024922, (2022) doi:10.1128/jvi.00249-22.
W. Zhang, K. Shi, Q. Geng, G. Ye, H. Aihara and F. Li, "Structural Basis for Mouse Receptor Recognition by SARS-CoV-2 Omicron Variant", Proc. Natl. Acad. Sci. USA 119, e2206509119 (2022) doi: 10.1073/pnas.2206509119

