Multiple sclerosis (MS) is a heterogeneous autoimmune disease in which autoreactive lymphocytes and antibodies attack the myelin sheath of the central nervous system (CNS). B lymphocytes in the cerebrospinal fluid (CSF) of MS patients contribute to inflammation and secrete oligoclonal immunoglobulins1,2. Epstein-Barr virus (EBV) infection has been linked to MS epidemiologically, but its pathological role remains unclear3,4.
Tobias Lanz et al.5 in William Robinson’s laboratory at Stanford University sequenced the single cell B cell repertoire in the cerebro-spinal fluid (CSF) and blood of MS patients in order to identify anti-viral and autoreactive antibodies. They selected clonally expanded antibody sequences, derived from the CSF of MS patients, and tested them for reactivity to viral and self-antigens using planar protein microarrays. Approximately one third of the selected antibodies bound to EBV proteins. The researchers identified one antibody (MS39p2w174), which binds with high affinity to the EBV transcription factor EBNA1 and cross-reacts to the glial cellular adhesion molecule GlialCAM. Interestingly, the unmutated germline ancestor of antibody MS39p2w174 binds EBNA1 with similar affinity as antibody MS39p2w174. In contrast, their binding affinities to GlialCAM differ significantly. While the affinity of MS39p2w174 to GlialCAM is ≥10-fold increased (KD EBNA1: 1.99 nM, SD: 0.63 nM; vs. KD GlialCAM: 190 pM, SD: 17pM), the germline antibody binds GlialCAM with lower affinity (KD EBNA1: 4.19 nM, SD: 0.76 nM vs. KD GlialCAM: 10.46 nM, SD: 4.12 nM).
The researchers inferred that a naïve B cell with an unmutated B cell receptor was activated by recognizing the EBV antigen EBNA1. It trafficked to the central nervous system and affinity matured towards increased affinity to the glial protein GlialCAM. Using mouse models of MS, the researchers showed that B cell reactivity against EBNA1 and GlialCAM aggravates neuroinflammation.
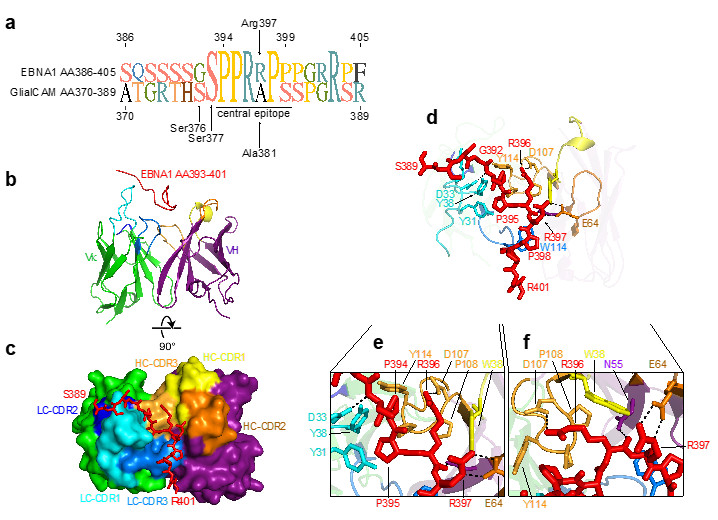
To understand the antigen-antibody interaction in detail, the researchers solved the structure of the EBNA1 peptide AA386-405 in complex with the Fab fragment of antibody MS39p2w174 at 2.5 Å resolution using macromolecular crystallography (PDB ID: 7K7R). The datasets were collected on the SSRL beam line 12-2.
The structure revealed close interactions of the EBNA1 residues P394-P398 with all CDRs except for light chain (LC) CDR2. Residues Tyr31 and Tyr38 on LC CDR1 together with Trp38 on heavy chain (HC) CDR1 and Pro108, Pro109, and Tyr114 on HC CDR3 create a hydrophobic cage for the peptide’s two N-terminal prolines Pro394 and Pro395 and the proximal side chain of Arg396 (Fig. 1a-j). The C-terminal end of the antibody binding groove is more open, and Pro398 is carried by a large aromatic tryptophan residue (Trp114 in LC CDR1) (Fig. 1h,j). The central arginines Arg395 and Arg396 hydrogen bond with residues on HC CDR2, HC CDR3, and HC framework region 2. Interestingly, all but two of the residues that directly interact with EBNA1 are unmutated germline residues.
Future projects will focus on understanding the molecular evolution of the described antibody from a naïve anti-viral antibody to a mutated self-reactive antibody. In addition to elucidating a mechanistic link between EBV and MS pathology the work promises to contribute to our fundamental understanding of the development of B cell self-reactivity on a molecular level and could be the basis for the development for antigen-specific therapies.
1. M. T. Cencioni, M. Mattoscio, R. Magliozzi, A. Bar-Or, and P. A. Muraro, "B Cells in Multiple Sclerosis - from Targeted Depletion to Immune Reconstitution Therapies", Nat. Rev. Neurol. 17, 399 (2021).
2. S. L. Hauser et al., "Ocrelizumab versus Interferon Beta-1a in Relapsing Multiple Sclerosis", N. Engl. J. Med. 376, 221 (2017).
3. A. Bar-Or et al., "Epstein–Barr Virus in Multiple Sclerosis: Theory and Emerging Immunotherapies", Trends Mol. Med. 26, 296 (2020).
4. K. Bjornevik et al., "Longitudinal Analysis Reveals High Prevalence of Epstein-Barr Virus Associated with Multiple Sclerosis", Science 375, 296 (2022).
5. Lanz, T. V. et al., "Clonally Expanded B Cells in Multiple Sclerosis Bind EBV EBNA1 and GlialCAM", Nature 603, 321–327 (2022).
T. V. Lanz, R. C. Brewer, P. P. Ho, J.-S. Moon, K. M. Jude, D. Fernandez, R. A. Fernandes, A. M. Gomez, G.-S. Nadj, C. M. Bartley, R. D. Schubert, I. A. Hawes, S. E. Vazquez, M. Iyer, J. B. Zuchero, B. Teegen, J. E. Dunn, C. B. Lock, L. B. Kipp, V. C. Cotham, B. M. Ueberheide, B. T. Aftab, M. S. Anderson, J. L. DeRisi, M. R. Wilson, R. J. M. Bashford-Rogers, M. Platten, K. C. Garcia, L. Steinman and W. H. Robinson, "Clonally Expanded B Cells in Multiple Sclerosis Bind EBV EBNA1 and GlialCAM", Nature 603, 321 (2022) doi: 10.1038/s41586-022-04432-7.

