Enzymes enable life on Earth by accelerating reactions with astounding efficiency and specificity (1, 2). How enzymes achieve their enormous rate enhancements has remained a central question in biology for over a century, and our understanding to date has extended our appreciation of evolutionary processes, influenced enzyme design, and impacted discovery and development of therapeutics.
The significant progress made in understanding enzyme chemical mechanisms has not been mirrored by progress in understanding the origins of enzyme catalytic efficiency. Enzymes bring together catalytic and substrate groups in active sites embedded within the folded enzyme and classic proposals to explain enzyme catalysis have invoked the importance of catalytic and substrate groups’ positioning for catalysis. While these proposals universally invoke positioning, the extent of positioning and the amount of catalysis provided by that positioning have been challenging to evaluate experimentally. Conversely, physics requires all atoms to undergo constant motion, forcing enzymes to exist as ensembles of conformational states and making it increasingly clear that conformational motions are also inherent to enzyme function.
A quantitative understanding of enzyme catalysis thus requires addressing these central questions: Are enzyme motions adjusted as the reaction proceeds? What is the extent of motion of active site catalytic residues? Are active site residues more precisely positioned than non-catalytic residues throughout an enzyme?
Enzyme conformational ensembles in which the extent of atomic motion is explicit can address fundamental questions about motions and positioning. While traditional structural biology approaches have been invaluable in biomedical research, they are limited in providing experimental conformational ensemble information: traditional x-ray crystallography models are only indirectly related to conformational ensembles; nuclear magnetic resonance experiments typically do not provide information about the absolute extent of motions; and molecular dynamics simulations do not report directly on actual physical behavior.
Researchers from Stanford University and the University of California at San Francisco in collaboration with SSRL obtained experimental conformation ensembles via two recently emerging x-ray crystallographic approaches: 1) “Pseudo-ensembles” obtained from a large number of high-sequence-similarity PDB models that when combined together capture ensemble behavior and 2) ensembles from x-ray data obtained near room temperature (3, 4). The researchers investigated the enzyme ketosteroid isomerase (KSI) which uses catalytic strategies common to many enzymes – an oxyanion hole and a general base mechanism (Fig. 1A). Focusing on KSI was essential to gaining both a general understanding of enzyme catalysis and answer some long-standing KSI-specific questions.
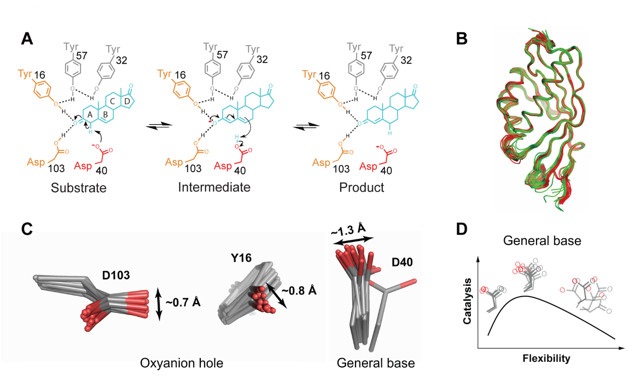
Figure 1. (A) The Ketosteroid Isomerase (KSI) reaction mechanism. KSI catalyzes double bond isomerization of steroid substrates utilizing a general base D40, and an oxyanion hole composed of the side chains of Y16 and D103. (B) Apo (green) and TSA-bound (red) KSI pseudo-ensembles. (C) The oxyanion hole (Y16 and D103) and general base (D40) conformational ensembles. (D) Conformational ensemble model for optimal general base catalysis. Optimal flexibility enables the general base to shuffle protons between different positions in different substrates (middle); reducing motion (left) reduces catalysis, as does increasing motion (right).
Pseudo-ensembles of KSI were constructed from apo and ligand-bound structures (Fig. 1B) and crystallographic datasets at elevated temperature were collected using SSRL beam line 9-2. The crystals were preserved in Paratone oil and diffraction data were collected at temperatures up to 280 K. Obtaining room temperature x-ray data was essential to eliminate potential artifacts that traditional cryo-cooling often introduces to the structure (5).
A proposed universal model for enzyme catalysis –that could not previously be tested– entailed gradual changes in the enzyme’s conformational ensemble upon binding of the substrate and in the transition state, such that the distribution of enzyme conformational states is most narrowed in the transition state, leading to a gradual reduction in conformational motion, with least enzyme motion in the transition state. However, analyses of KSI ensembles that capture the different states of the enzyme’s reaction cycle indicated only modest decrease in enzyme conformational motion upon substrate binding, and no further decrease in motion upon transitioning from the substrate-bound to the transition state.
Classic proposals for enzyme catalysis have also invoked the importance of positioning and restricting the motions of active site groups, but the extent to which these motions are restricted was previously unknown for any enzyme. The researchers’ analyses of the KSI conformational ensembles indicated that both oxyanion hole and general base catalytic residues exhibit motions of 1 – 1.5 Å - the scale of a bond length (Fig. 1C). To evaluate if the motions of the catalytic residues are exceptionally well positioned, the researchers defined the metric “mean deviation” (MDev) that quantifies the width of an ensemble of atoms, with a narrow ensemble giving low MDev values while a wider ensemble – high MDev values. The catalytic groups’ MDev values were at the lower end but not lower than MDev values for similar non-catalytic groups, suggesting no exceptional or extraordinary precision in the positioning of the catalytic groups at the active site of KSI.
The exact mechanism rendering the general base, a highly efficient catalytic feature in the context of KSI, was previously unknown (6). The KSI conformational ensembles allowed the researchers to test models that account for this catalytic efficiency. The KSI general base shuffles protons between different positions in substrates. Because the above results indicated a general base motion on the scale of 1 – 1.5 Å, the researchers hypothesized that increasing or decreasing the extent of this motion would decrease the general base catalytic efficiency. To test this hypothesis, they departed from the traditional “structure-function” relationships and introduced “ensemble-function” relationships obtained by KSI ensembles in which the extent of general base motion was increased or decreased compared to wild type, and they combined these with functional data. These studies indicated that increased general base motion appears to be required for function but that there is an optimal balance between allowing and limiting motion for efficient catalysis (Fig. 1D).
- J. M. Berg, et al., Biochemistry, 5th Ed. (W H Freeman, 2002).
- R. Wolfenden, M. J. Snider, "The Depth of Chemical Time and the Power of Enzymes as Catalysts", Acc. Chem. Res. 34, 938 (2001).
- R. B. Best, K. Lindorff-Larsen, M. A. DePristo, M. Vendruscolo, "Relation between Native Ensembles and Experimental Structures of Proteins", Proc. Natl. Acad. Sci. USA 103, 10901 (2006).
- H. van den Bedem, A. Dhanik, J.-C. Latombe, A. M. Deacon, "Modeling Discrete Heterogeneity in X-ray Diffraction Data by Fitting Multi-conformers. Acta Crystallogr. D Biol. Crystallogr. 65, 1107 (2009).
- B. Halle, Biomolecular Cryocrystallography: Structural Changes during Flash-cooling", Proc. Natl. Acad. Sci. USA 101, 4793 (2004).
- V. Lamba, F. Yabukarski, M. Pinney, D. Herschlag, Evaluation of the Catalytic Contribution from a Positioned General Base in Ketosteroid Isomerase. J. Am. Chem. Soc. 138, 9902 (2016).
F. Yabukarski, J. T. Biel, M. M. Pinney, T. Doukov, A. S. Powers, J. S. Fraser and D. Herschlag, "Assessment of Enzyme Active Site Positioning and Tests of Catalytic Mechanisms through X-ray–derived Conformational Ensembles", Proc. Natl. Acad. Sci. USA 29, 33204 (2020) doi: 10.1073/pnas.2011350117

