Monoclonal antibodies are valuable weapons in the battle against COVID-19 as direct-acting antiviral agents (1). Central to virus replication cycle, the SARS-CoV-2 spike protein binds the host cell receptor and engages in virus-host membrane fusion (2). Conformational flexibility of the spike protein allows each of its receptor binding domains (RBDs) to exist in two major configurations: a “down” conformation that is thought to be less accessible to binding of many neutralizing antibodies and an “up” conformation that binds both the receptor and neutralizing antibodies (3-5). Some neutralizing antibodies bind to the RBD in the “up” conformation and compete with the receptor (6, 7), while some neutralizing antibodies bind and stabilize the “down” conformation to prevent the conformational changes required for viral entry, thereby hindering infection (8, 9).
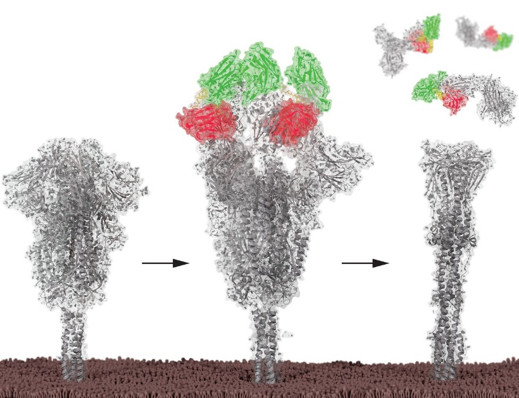
Figure 1. Bivalent nanobodies inducing post-fusion conformation of the SARS-CoV-2 spike protein: SARS-CoV-2 spike proteins are in a fusion inactive configuration when the RBDs are in the down conformation (left). Binding of bivalent nanobody (red and green ribbons joined by yellow tether) stabilizes the spike in an active conformation with all RBDs up (middle), triggering premature induction of the post-fusion conformation, which irreversibly inactivates the spike protein (right).
Unfortunately, antibody molecules can be more difficult to produce in large quantities and are relatively costly to produce. Single domain antibodies, also known as nanobodies, offer an opportunity to rapidly produce antiviral agents for immunization and for therapy. Nanobodies are easier to produce, have high thermal stability and have the potential to be administered by inhalation.
A consortium of researchers from the University of Bonn, The Scripps Research Institute, the Karolinska Institutet and the University of Illinois at Urbana-Champagne designed, engineered, and structurally characterized new nanobodies. They used the resulting detailed structural information of the nanobody binding modes to optimize for a more effective COVID-19 therapeutic candidate.
Nanobodies specific for the receptor binding domain (RBD) of the SARS-CoV-2 spike protein were identified by phage display using nanobody libraries from an alpaca and a llama immunized with the RBD and inactivated virus. X-ray crystallography performed on SSRL BL12-1, as well as at the APS, of four resulting nanobodies revealed that they bind to distinct epitopes on the RBD. Based on this structural information, the authors were able to design bi- and tri-valent nanobodies with improved neutralizing properties. One of the multivalent nanobodies that simultaneously targeted two independent epitopes on the spike protein was able to activate the fusion machinery (Fig. 1). Activation of spike in the absence of target membranes induces irreversible conformational changes without promoting fusion, thus preventing cell entry and virus replication. These engineered multivalent nanobodies provided more than 100 times the neutralizing activity of monovalent nanobodies and have strong potential clinical applications, owing to the increased neutralization activity and the built-in protection from rapid emergence of escape mutants.
- J. Abraham, "Passive Antibody Therapy in COVID-19", Nat. Rev. Immunol. 20, 401 (2020).
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Müller, C. Drosten, and S. Pöhlmann, "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and is Blocked by a Clinically Proven Protease Inhibitor", Cell 181, 271 (2020).
- D. Wrapp, N. Wang, K. S. Corbett, J. A. Goldsmith, C. L. Hsieh, O. Abiona, B. S. Graham, and J. S. McLellan, "Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation", Science 367, 1260 (2020).
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire, and D. Veesler, "Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein", Cell 181, 281 (2020).
- Z. Lv, Y.-Q. Q. Deng, Q. Ye, L. Cao, C.-Y. Y. Sun, C. Fan, W. Huang, S. Sun, Y. Sun, L. Zhu, Q. Chen, N. Wang, J. Nie, Z. Cui, D. Zhu, N. Shaw, X.-F. F. Li, Q. Li, L. Xie, Y. Wang, Z. Rao, C.-F. F. Qin, and X. Wang, "Structural Basis for Neutralization of SARS-CoV-2 and SARS-CoV by a Potent Therapeutic Antibody", Science 369, 1505 (2020).
- T. F. Rogers, F. Zhao, D. Huang, N. Beutler, A. Burns, W. T. He, O. Limbo, C. Smith, G. Song, J. Woehl, L. Yang, R. K. Abbott, S. Callaghan, E. Garcia, J. Hurtado, M. Parren, L. Peng, S. Ramirez, J. Ricketts, M. J. Ricciardi, S. A. Rawlings, N. C. Wu, M. Yuan, D. M. Smith, D. Nemazee, J. R. Teijaro, J. E. Voss, I. A. Wilson, R. Andrabi, B. Briney, E. Landais, D. Sok, J. G. Jardine, and D. R. Burton, "Isolation of Potent SARS-CoV-2 Neutralizing Antibodies and Protection from Disease in a Small Animal Model", Science 369, 956 (2020).
- C. O. Barnes, A. P. West Jr.., K. E. Huey-Tubman, M. A. G. Hoffmann, N. G. Sharaf, P. R. Hoffman, N. Koranda, H. B. Gristick, C. Gaebler, F. Muecksch, J. C. C. Lorenzi, S. Finkin, T. Hägglöf, A. Hurley, K. G. Millard, Y. Weisblum, F. Schmidt, T. Hatziioannou, P. D. Bieniasz, M. Caskey, D. F. Robbiani, M. C. Nussenzweig, and P. J. Bjorkman, "Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies", Cell 182, 828 (2020).
- M. Schoof, B. Faust, R. A. Saunders, S. Sangwan, V. Rezelj, N. Hoppe, M. Boone, C. B. Billesbølle, C. Puchades, C. M. Azumaya, H. T. Kratochvil, M. Zimanyi, I. Deshpande, J. Liang, S. Dickinson, H. C. Nguyen, C. M. Chio, G. E. Merz, M. C. Thompson, D. Diwanji, K. Schaefer, A. A. Anand, N. Dobzinski, B. S. Zha, C. R. Simoneau, K. Leon, K. M. White, U. S. Chio, M. Gupta, M. Jin, F. Li, Y. Liu, K. Zhang, D. Bulkley, M. Sun, A. M. Smith, A. N. Rizo, F. Moss, A. F. Brilot, S. Pourmal, R. Trenker, T. Pospiech, S. Gupta, B. Barsi-Rhyne, V. Belyy, A. W. Barile-Hill, S. Nock, Y. Liu, N. J. Krogan, C. Y. Ralston, D. L. Swaney, A. García-Sastre, M. Ott, M. Vignuzzi, P. Walter, and A. Manglik, "QCRG Structural Biology Consortium, An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike", Science 370, 1473 (2020).
- M. A. Tortorici, M. Beltramello, F. A. Lempp, D. Pinto, H. V. Dang, L. E. Rosen, M. McCallum, J. Bowen, A. Minola, S. Jaconi, F. Zatta, A. De Marco, B. Guarino, S. Bianchi, E. J. Lauron, H. Tucker, J. Zhou, A. Peter, C. Havenar-Daughton, J. A. Wojcechowskyj, J. B. Case, R. E. Chen, H. Kaiser, M. Montiel-Ruiz, M. Meury, N. Czudnochowski, R. Spreafico, J. Dillen, C. Ng, N. Sprugasci, K. Culap, F. Benigni, R. Abdelnabi, S. C. Foo, M. A. Schmid, E. Cameroni, A. Riva, A. Gabrieli, M. Galli, M. S. Pizzuto, J. Neyts, M. S. Diamond, H. W. Virgin, G. Snell, D. Corti, K. Fink, and D. Veesler, "Ultrapotent Human Antibodies Protect against SARS-CoV-2 Challenge via Multiple Mechanisms", Science 370, 950 (2020).
P.-A. Koenig, H. Das, H. Liu, B. M. Kummerer, F. N. Gohr, L.-M. Jenster, L. D. J. Schiffelers, Y. M. Tesfamariam, M. Uchima, J. D. Wuerth, K. Gatterdam, N. Ruetalo, M. H. Christensen, C. I. Fandrey, S. Normann, J. M. P. Todtmann, S. Pritzl, L. Hanke, J. Boos, M. Yuan, X. Zhu, J. L. Schmid-Burgk, H. Kato, M. Schindler, I. A. Wilson, M. Geyer, K. U. Ludwig, B. M. Hallberg, N. C. Wu and F. I. Schmidt, "Structure-guided Multivalent Nanobodies Block SARS-CoV-2 Infection and Suppress Mutational Escape", Science 371, eabe6230 (2021) doi: 10.1126/science.abe6230

