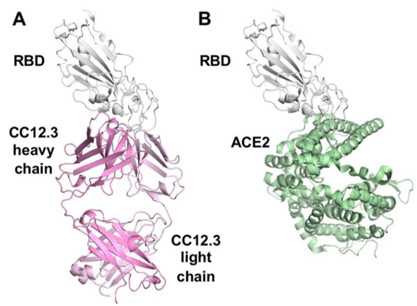
Figure 1. Binding of the SARS-CoV-2 receptor binding domain (RBD) (gray ribbons) to (A) the neutralizing antibody (pink ribbons) and to (B) the ACE2 human cell surface receptor (green). The antibody binds to the same site as the human receptor as suggested by competition studies, and would be expected to effectively block the binding of the virus to the cell and prevent viral entry.
The COVID-19 pandemic caused by the SARS-CoV-2 coronavirus is causing health and economic havoc on a global scale requiring the development of an effective vaccine and therapeutics. Spike (S) proteins found on the viral surface are the major surface antigens of SARS-CoV-2. The S protein uses its receptor-binding domain (RBD) to attach the host receptor ACE2 for viral entry into the cell.1 Neutralizing antibodies which target the RBD of the CoV-2 virus would effectively block viral entry. More than 290 antibodies that target the RBD of SARS-CoV-2 were analyzed, and the immunoglobulin heavy chain gene IGHV3-53 was the most frequently used to produce these antibodies.2 These IGHV3-53 antibodies not only had lower mutation rates, but were also more potent compared to other germlines. Prior research suggests that antibodies encoded by IGHV3-53 are generally present, at least in small numbers, in healthy people’s blood.3
A research team led by Ian Wilson at The Scripps Research Institute used X-ray crystallography to determine the structures of two of these antibodies (CC12 .1 and CC12.3) from convalescent patients in complex with the SARS-CoV-2 RBD. Diffraction data were collected at the newly-commissioned high-brilliance micro-focus SSRL beam line 12-1. The structures showed that the CC12.1 and CC12.3 antibodies bound to the same site on the RBD, a site already identified as the ACE2 binding site and ACE2 binding residues on RBD were found to be within the binding epitopes of the antibodies. The fact that the antibodies and the target receptor both bind to the same site on the viral RBD explains the strong neutralizing activity of these IGHV3-53-encoded antibodies. The structures revealed unique features of the antibodies that facilitate their high binding affinity and their specificity for the SARS-CoV-2 RBD.
The detailed atomic-scale structure of the antibody interactions with the virus should help accelerate vaccine design. Moreover the identification of IGHV3-53-encoded antibodies as key elements of the immune response to COVID-19 suggests that levels of these antibodies might be useful as an indirect marker of success in ongoing and future vaccine trials.
1. R. Yan, Y. Zhang, Y. Li, L. Xia, Y. Guo and Q. Zhou, "Structural Basis for the Recognition of SARS-CoV-2 by Full-length Human ACE2", Science 367, 1444 (2020)
2. M. Yuan, N. C. Wu, X. Zhu, C. D. Lee, R. T. Y. So, H. Lv, C. K. P. Mok and I. A. Wilson, "A Highly Conserved Cryptic Epitope in the Receptor Binding Domains of SARS-CoV-2 and SARS-CoV", Science 368, 630 (2020)
3. B. Briney, A. Inderbitzin, C. Joyce and D. R. Burton, "Commonality Despite Exceptional Diversity in the Baseline Human Antibody repertoire", Nature 566, 393 (2019)
M. Yuan, H. Liu, N. C. Wu, C.-C. D. Lee, X. Zhu, F. Zhao, D. Huang, W. Yu, Y. Hua, H. Tien, T. F. Rogers, E. Landais, D. Sok, J. G. Jardine, D. R. Burton and I. A. Wilson, "Structural Basis of a Shared Antibody Response to SARS-CoV-2", Science 369, 1119 (2020) DOI: 10.1126/science.abd2321

