Life on Earth is dictated by the DNA alphabet comprised of only four DNA bases or letters: A, T, G and C. It has long been of interest to understand whether there is something very special about the four letters that comprise DNA and whether this is the only code that could support life. At a basic level, this question can be addressed by examining an expanded alphabet and determining the properties of DNA including additional synthetic letters. This study impacts our current understanding of terrestrial DNA and suggests that extraterrestrial life forms could have evolved using a different genetic code than found here on Earth. The work has immediate applications in synthetic biology for the creation of new molecules and greatly expands the ability to store information in DNA.
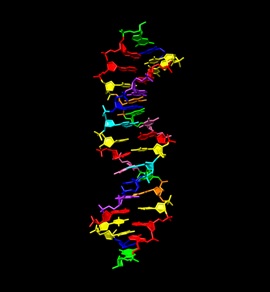
Figure. Crystal structure of a double helix built from eight hachimoji building blocks, G (green), A (red), C (dark blue), T (yellow), B (cyan), S (pink), P (purple), and Z (orange). The first four building blocks are found in human DNA; the last four are synthetic, and possibly present in alien life. Each strand of the double helix has the sequence CTTAPCBTASGZTAAG. Notable is the geometric regularity of the pairs, a regularity that is needed for evolution.
Now, in breakthrough work, funded by NASA, NSF and NIGMS, Dr. Steven Benner at the Foundation for Applied Molecular Evolution, in collaboration with Dr. Millie Georgiadis at the Indiana University School of Medicine, and colleagues at biotechnology companies and other universities, have provided evidence that the standard DNA code can be expanded to include eight letters forming “hachimoji DNA” (“hachi” eight and “moji” letter in Japanese) using four novel synthetic nucleobases (B, S, P and Z) in addition to A, T, C and G and still retain critical features of natural DNA1,2. Structurally, hachimoji DNA can adopt a standard double helical form of DNA and retain Watson-Crick complementary base pairing, which allows the expanded DNA to be faithfully replicated and transcribed by polymerases to produce hachimoji DNA copies and hachimoji RNA. These properties are essential for a genetic system that can support life.
The researchers used SSRL Beam Line 14-1 for macromolecular crystallography experiments of their hachimoji DNA, which included the novel bases B, S, P and Z along with the natural bases A, C, G, and T. To obtain a view of the structural properties of the DNA without issues of distortion from the crystallization lattice, they used a clever host-guest complex system, in which the hachimoji DNA is co-crystallized with part of a reverse transcriptase protein. These structural studies confirmed that hachimoji DNA forms a regular DNA helix. In complementary biochemical experiments, they found that DNA polymerase and RNA polymerase recognize this DNA and can correctly copy the new bases, showing that the synthetic bases have the same functionality as the natural bases.
Creation of these new DNA letters and duplex structures involved several steps. Interaction thermodynamics of these DNA letters was assessed by synthesizing DNA using solid-phase chemistry and measuring thermodynamic data for base pair dimers. This information was then used to generate new base pair dimer parameters using singular value decomposition methods. Experimental and predicted DNA melting curves were then measured and calculated for validation and comparison and it was concluded that the expanded DNA code GACTZPSB captures the molecular recognition behavior of standard four-letter DNA, thus allowing for an expanded system of information storage.
Experimental validation of the structural properties of hachimoji DNA was achieved through the use of a host-guest complex system, developed by the Georgiadis group, which facilitates rapid crystallization, structure determination, and analysis of novel DNA sequences by macromolecular crystallography. In this system, the N-terminal portion of a reverse transcriptase is used as a host to assemble crystals of interesting DNA sequences forming protein-DNA crystals in which the inherent properties of the DNA structure can be studied independent of DNA lattice packing effects. The structure determinations were expedited by Accelero Biostructures, a San Francisco-based protein crystallography-driven drug discovery company cofounded by Dr. Debanu Das, a former graduate student of Dr. Georgiadis, and Dr. Ashley Deacon, who provided rapid, high-quality macromolecular crystallography data collection and processing on crystals of the protein-DNA complexes using the SSRL Beam Line 14-1. The crystal structures provided 3D proof that hachimoji DNA assembles duplex DNA retaining essential features of natural DNA while imparting novel sequence specific features conferred by the novel synthetic base pairs.
- M. M. Georgiadis, I. Singh, W. F. Kellett, S. Oshika, S. A. Benner and N. G. J. Richards, “Structural Basis for a Six Nucleotide Genetic Alphabet”, J. Am. Chem. Soc. 137, 6947 (2015).
- S. Hoshika, I. Singh, C. Switzer, R. W. Molt Jr., N. A. Leal, M. J. Kim, M. S. Kim, H. J. Kim, M. M. Georgiadis and S. A. Benner, "Skinny" and "Fat" DNA: Two New Double Helices”, J. Am. Chem. Soc. 140, 11655 (2018).
S. Hoshika, N. A. Leal, M.-J. Kim, M.-S. Kim, N. B. Karalkar, H.-J. Kim, A. M. Bates, N. E. Watkins, Jr., H. A. SantaLucia, A. J. Meyer, S. DasGupta, J. A. Piccirilli, A. D. Ellington, J. J. SantaLucia, M. M. Georgiadis and S. A. Benner, "Hachimoji DNA and RNA: A Genetic System with Eight Building Blocks", Science 363, 884 (2019) doi: 10.1126/science.aat0971

