
Figure 1. Cartoon representation of Ago2 showing the location of the tryptophan-binding region.
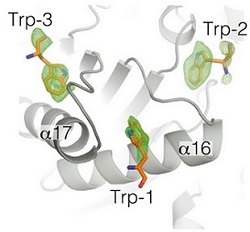
Figure 2. Close up view of the trp-binding region. Fo-Fc tryptophan omit map, contoured at 2.5 σ, (green mesh) shows well-ordered indole side chains of three bound tryptophan molecules.
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that are integral to diverse physiological processes in animals, including tissue regeneration, cardiac function, brain development, and the progression of cancer. To exert their regulatory function, miRNAs associate with Argonaute proteins to form the core of the miRNA-induced silencing complex (miRISC). miRISC is a multi-protein assembly that uses miRNAs to identify mRNAs targeted for repression and uses the sequence information in the miRNA as a guide to bind complementary sequences in mRNAs targeted for silencing. Once bound to its target mRNA, miRISC induces silencing by elements of both translational repression and mRNA decay. miRISC appears to be able to silence from any position on an mRNA to which it can stably bind. Dozens of miRISC-associated proteins have been identified, and interactions between many factors have been examined in detail. However, the physical nature of the complex remains unknown.
In this study, Drs. Ian MacRae and Jessica Sheu-Gruttadauria used purified samples of two core human miRISC components, Argonaute2 (Ago2) and TNRC6B, to examine the assembly, activity, and physical properties of reconstituted human miRISC. Ago2 was known to bind to tryptophan residues in the glycine-tryptophan rich domain in TNRC6B. A 2.2 Å diffraction data set of Ago2, crystallized in the presence of tryptophan (trp), was collected using remote-access at Beam Line 12-2. The results allowed the identification of three evenly spaced trp-binding pockets clustered together in a small region near the bottom of the Ago2 PIWI domain. The trp-binding pockets are arranged in an equilateral triangle on the surface of Ago2, and Ago2 only contacts the indole side chains of bound trp molecules, leaving the main chain atoms free to rotate about the Cα-Cβ bond. (Figure 2). Thus, appropriately spaced trp residues in TNRC6B could potentially interact with any two trp-binding pockets, thereby allowing the formation of complex multivalent interactions between the two proteins. These multivalent interactions were then shown to promote condensation of Ago2 and TNRC6B into phase-separated droplets in vitro and in live cells. Phase separation of the core miRISC components was shown to accelerate mRNA decay in vitro, and proposed to be used by cells to regulate gene expression by controlling the stability and localization of mRNAs targeted by miRNAs.
The miRISC droplets formed in vitro recruit deadenylation factors and sequester target RNAs from the bulk solution. The condensation of miRISC is accompanied by accelerated deadenylation of target RNAs bound to Ago2. The combined results may explain how miRISC silences mRNAs of varying size and structure and provide experimental evidence that protein-mediated phase separation can facilitate an RNA processing reaction.
The authors hypothesize that the ability to form higher order complexes via molecular condensation may allow miRISC to organize miRNA-targets within the cytoplasm and thereby modulate rates of mRNA translation and decay.
J. Sheu-Gruttadauria and I. J. MacRae, "Phase Transitions in the Assembly and Function of Human miRISC", Cell 173, 946 (2018) doi: 10.1016/j.cell.2018.02.051

