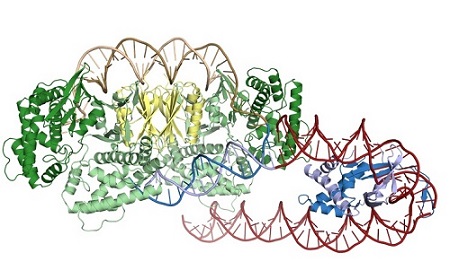
Structure of Cas1-Cas2 (green and yellow) bound to a half-site intermediate. IHF (blue) bends the leader DNA (red) to allow for an additional sequence-specific interaction with Cas1.
CRISPR (Clustered regularly-interspaced short palindromic repeats) immune systems provide prokaryotes with adaptive immunity against phage and other foreign genetic elements. The first stage of immunity, acquisition, entails the capture of foreign DNA and its subsequent insertion at the CRISPR locus, which serves as a repository of viral sequences to allow for identification upon reinfection1. The integration of new sequences is carried about by Cas1-Cas2, the most conserved elements of the otherwise diverse CRISPR systems2. Cas1-Cas2 must restrict their activity to the CRISPR locus to ensure that captured viral DNA leads to productive immunity and avoid introducing mutations through the off-target integration of viral sequences. Integration occurs specifically at the first of a series of direct repeats, that, together with the intervening viral-derived spacers, comprise the CRISPR array. The critical sequences within the repeat and the adjacent leader region have been identified, but the mechanism by which Cas1-Cas2 recognize these sequences is unknown3,4.
New work by Jennifer Doudna’s group at the University of California, Berkeley, reveals that the Cas1-Cas2 from the Escherichia coli CRISPR system rely heavily on indirect sequence recognition, rather than direct readout of the repeat sequence, to restrict their activity to the CRISPR locus5. The group captured Cas1-Cas2 bound to substrates representing both a partially-integrated and a fully-integrated fragment of DNA. Using x-ray diffraction data sets collected at SSRL BL9-2 and ALS 8.3.1, they solved the structures of the half-site intermediate and full-site product complexes. The structures revealed a striking lack of sequence-specific contacts between Cas1-Cas2 and the repeat DNA. Instead, the repeat DNA was found to be highly distorted. For both ends of the repeat to be captured by the two catalytic Cas1 active sites, the DNA had to both bend and unwind as it traversed the complex, and biochemical experiments suggest that the repeat sequence itself confers the required flexibility. The group also solved the structure of the complex in the presence of IHF (integration host factor), a DNA-bending protein required for recognition of the leader, using cryo-electron microscopy. IHF introduces a 180° bend in the leader, allowing Cas1 to interact with an upstream recognition motif6.
The native information storage activity of Cas1-Cas2 can be repurposed for a variety of functions, such as barcoding individual cells, encoding information about cellular states, or, as has recently been shown, recording a movie in E. coli genomes7. Many of these applications would require redirecting Cas1-Cas2 to recognize a non-cognate sequence. The insights about the mode of target recognition provided by the structures can inform both the prediction of potential recognition sites in other genomes and the engineering of Cas1-Cas2 to recognize a sequence of choice, allowing for Cas1-Cas2 to be used outside of their native context.
- A. V. Wright, J. K. Nuñez and J. A. Doudna, "Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering", Cell 164, 29 (2016) doi: 10.1016/j.cell.2015.12.035.
- K. S. Makarova et al., "An Updated Evolutionary Classification of CRISPR–Cas Systems", Nat. Rev. Microbiol. 13, 722 (2015). doi:10.1038/nrmicro3569
- M. G. Goren et al., "Repeat Size Determination by Two Molecular Rulers in the Type I-E CRISPR Array", Cell Rep. 16, 2811 (2016).
- C. Moch, M. Fromant, S. Blanquet and P. Plateau, "DNA Binding Specificities of Escherichia coli Cas1–Cas2 Integrase Drive Its Recruitment at the CRISPR Locus", Nucleic Acids Res. 45, 2714 (2016) doi:10.1093/nar/gkw1309.
- A. V. Wright et al., "Structures of the CRISPR Genome Integration Complex", Science 64, eaao0679 (2017) doi: 10.1126/science.aao0679.
- K. N. R. Yoganand, R. Sivathanu, S. Nimkar and B. Anand, "Asymmetric Positioning of Cas1–2 Complex and Integration Host Factor Induced DNA Bending Guide the Unidirectional Homing of Protospacer in CRISPR-Cas Type I-E System", Nucleic Acids Res. 45, 367 (2017) doi: 10.1093/nar/gkw1151.
- S. L. Shipman, J. Nivala, J. D. Macklis and G. M. Church, "CRISPR–Cas Encoding of a Digital Movie into the Genomes of a Population of Living Bacteria. Nature 547, 345 (2017) doi:10.1038/nature23017.
A. V. Wright, J.-J. Liu, G. J. Knott, K. W. Doxzen, E. Nogales and J. A. Doudna, "Structures of the CRISPR Genome Integration Complex", Science 64, eaao0679 (2017) doi: 10.1126/science.aao0679.

