Pili (or fimbriae) are hair-like structures on the cell surface of many bacteria. Pili can play a variety of functions in bacteria, such as conjugation, mobility and adhesion, and often serve as primary virulence factors. Pili are formed by noncovalent or covalent oligomerization of component proteins, pilins or fimbrilins. For example, the type I pilus of Escherichia coli is assembled through exchange of β-strands between pilins. Gram-negative Porphyromonas gingivalis, a major oral pathogen associated with severe adult periodontitis and other diseases, produces two types of pili (major and minor) that are key virulence factors essential for host colonization, evasion of innate defenses, and biofilm formation. However, P. gingivalis pilins have no similarity to other characterized pilins. Using x-ray macromolecular crystallography data collected from SSRL and ALS beam lines, researchers from the Joint Center for Structural Genomics (JCSG) determined 20 structures of P. gingivalis pilins or their homologs from the human gut microbiome. These structures, together with comprehensive biochemical analysis in collaboration with the Nakayama group at Nagasaki University in Japan, provided deep insights into the evolutionary origin, assembly mechanism, and distribution of this new type of pilus (type V).
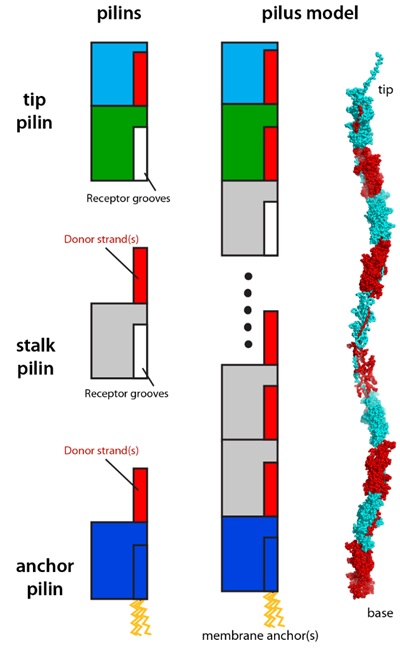
All type V pre-pilins contain a common two-domain fold that serves as the assembly module. Each β-sandwich domain contains seven core strands, and adopt a topology that is approximately inverse to that of the IgG fold. During the assembly process, the first core strand of the N-terminal domain is cleaved by a protease, resulting in an incomplete six-strand fold with an exposed surface groove, which is complemented by extra strands from the C-terminal domain of the incoming pilin. The polymerization process is terminated by a specialized anchor pilin that does not possess an N-terminal domain receptor groove, since its first strand is recalcitrant to protease cleavage. The pilin at the tip of pilus may possess extra functional modules, such as lectin domains that may facilitate binding of surface polysaccharides of target cells. Overall, type V pilus shares a similar strand-exchange assembly mechanism to that of the type I pilus, despite its pilins having a fold similar to that of the Gram-positive pilins that polymerize via covalent bonds.
Type V pilin-related proteins are highly abundant in bacterial species in the gut microbiome and are extremely divergent. As such, they adopt a common structural scaffold but allow for recognition of different binding partners or evolution of new functional roles, and may represent an important class of molecules that are involved in both symbiotic and pathogenesis relationships between the human host and its microbiome. The current study provides a foundation for further exploration of these molecules in host microbe interactions, structure-based drug design, and vaccine development.
- W. J. Allen, G. Phan and G. Waksman, "Pilus Biogenesis at the Outer Membrane of Gram-negative Bacterial Pathogens", Curr. Opin. Struct. Biol. 22, 500 (2012).
- T. Proft and E. N. Baker, "Pili in Gram-negative and Gram-positive Bacteria - Structure, Assembly and Their Role in Disease", Cell Mol. Life Sci. 66, 613 (2009).
- Q. Xu, P. Abdubek, T. Astakhova, H. L. Axelrod, C. Bakolitsa, X. Cai, D. Carlton, C. Chen, H.-J. Chiu, M. Chiu, T. Clayton, D. Das, M. C. Deller, L. Duan, K. Ellrott, C. L. Farr, J. Feuerhelm, J. C. Grant, A. Grzechnik, G. W. Han, L. Jaroszewski, K. K. Jin, H. E. Klock, M. W. Knuth, P. Kozbial, S. S. Krishna, A. Kumar, D. Marciano, D. McMullan, M. D. Miller, A. T. Morse, E. Nigoghossian, A. Nopakun, L. Okach, C. Puckett, R. Reyes, N. Sefcovic, H. J. Tien, C. B. Trame, H. van den Bedem, D. Weekes, T. Wooten, A. Yeh, J. Zhou, K. O. Hodgson, J. Wooley, M.-A. Elsliger, A. M. Deacon, A. Godzik, S. A. Lesley and I. A. Wilson, "A Conserved Fold for Fimbrial Components Revealed by the Crystal Structure of a Putative Fimbrial Assembly Protein (BT1062) from Bacteroides thetaiotaomicron at 2.2 Å Resolution", Acta Crystallogr. F 66, 1281 (2010), DOI: 10.1107/S1744309110006548.
- Q. Xu, M. Shoji, S. Shibata, M. Naito, K. Sato, M.-A. Elsliger, J. C. Grant, H. L. Axelrod, H.-J. Chiu, C. L. Farr, L. Jaroszewski, M. W. Knuth, A. M. Deacon, A. Godzik, S. A. Lesley, M. A. Curtis, K. Nakayama and I. A. Wilson, "A Distinct Type of Pilus from the Human Microbiome", Cell 165, 690 (2016), DOI: 10.1016/j.cell.2016.03.016.

