Scientists in the Structural Molecular Biology (SMB) program at the Stanford Synchrotron Radiation Lightsource (SSRL) in collaboration with scientists at Stanford University and at the Linac Coherent Light Source (LCLS) developed a goniometer-based system to study radiation-sensitive macromolecular complexes. The system operates in air and is complementary to the injector-based in-vacuum crystallographic x-ray diffraction experiments traditionally performed with the LCLS X-ray Free Electron Laser (XFEL). Both methods take advantage of the “diffraction before destruction” approach (x-ray diffraction images are recorded before the crystal sample is destroyed). This is achieved using the characteristics of the XFEL which provides a highly intense x-ray pulse that is extremely short - only 10’s of femtoseconds in duration - thus “beating out” damage from radiation and heat.
Injector-based experiments that are typically done at the LCLS, inject millions of tiny, randomly oriented crystals in a thin liquid stream into the path of the XFEL pulses. Many crystals pass by between x-ray pulses and are not exposed. In contrast, the goniometer-based system centers one crystal at a time into each x-ray pulse. This greatly reduces the number of crystals needed for structural studies on rare and important samples that require a more controlled approach. This system is also ideal for studying larger crystals which may contain enzymes that are extremely radiation sensitive.
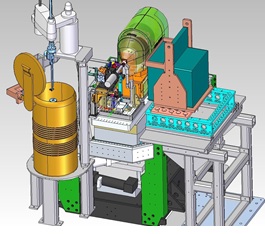
Figure 1. A schematic representation of the SMB goniometer-based setup as currently installed on the XPP station at the LCLS.
The new system, figure 1, has been used exclusively at the X-ray Pump Probe (XPP) station at LCLS and includes a high-speed high-precision goniometer developed by the SMB group for the microfocus station BL12-2 at SSRL. It also incorporates a sample mounting robot and options for studying cryogenically-cooled or room-temperature samples. Large-area CCD and rapid-readout PAD detectors are available for data collection. The new setup is described in the Proceedings of the National Academy of Sciences [1].
The system has already been used to provide a complete picture of an enzyme structure in about 30 minutes using only five crystals, using a helical mode of data collection where long crystals were translated and rotated between diffraction “snapshots.” It was also used to determine the structure of a protein complex that controls the release of signaling chemicals called neurotransmitters from brain cells [2] (see next highlight). In the latter case, numerous individual frozen crystals were mounted with a robot and a “point-and-click” approach was used to move samples into the beam and to collect diffraction images.
A new beam line called the Macromolecular Femtosecond Crystallography (MFX) station is currently under construction at LCLS. It will be dedicated to house the goniometer-based experimental equipment that has traditionally been installed and removed each time it is used at the XPP station. The new beam line may also generate additional beam time by making use of x-ray pulses that become available when they are not being used during other experiments (for example during sample exchanges). X-ray pulses would be temporarily directed to MFX using a deflecting mirror. Goniometer-based experiments will continue to be scheduled on XPP until the MFX station is fully operational in 2016.
1. A. E. Cohen, et al., Proc. Natl. Acad. Sci. USA 111, 17122 (2014).
2. Q. Zhou, et al., Nature 525, 62 (2015).
A. E. Cohen, S. M. Soltis, A. González, L. Aguila, R. Alonso-Mori, C. O. Barnes, E. L. Baxter, W. Brehmer, A. S. Brewster, A. T. Brunger, G. Calero, J. F. Chang, M. Chollet, P. Ehrensberger, T. L. Eriksson, Y. Feng, J. Hattne, B. Hedman, M. Hollenbeck, J. M. Holton, S. Keable, B. K. Kobilka, E. G. Kovaleva, A. C. Kruse, H. T. Lemke, G. Lin, A. Y. Lyubimov, A. Manglik, I. I. Mathews, S. E. McPhillips, S. Nelson, J. W. Peters, N. K. Sauter, C. A. Smith, J. Song, H. P. Stevenson, Y. Tsai, M. Uervirojnangkoorn, V. Vinetsky, S. Wakatsuki, W. I. Weis, O. A. Zadvornyy, O. B. Zeldin, D. Zhu and K. O. Hodgson, "Goniometer-based Femtosecond Crystallography with X-ray Free Electron Lasers", Proc. Natl. Acad. Sci. USA 111, 17122 (2014), DOI: 10.1073/pnas.1418.

