The Escherichia coli (E. coli) proteome consists of 5993 proteins, of which 853 are involved in primary metabolic processes critical for the survival and functioning of the cell1. Fatty acid biosynthesis is at the core of primary metabolism responsible for the synthesis of fatty acids, essential metabolites that are the major components of cellular membranes and energy storage. Due to the high prevalence of multi-antibiotic resistant bacteria, successful models for the inhibition of fatty acid biosynthesis enable novel antibiotics developments. Conversely, an efficiently tailored production of fatty acids promises “green” solutions to the current energy crisis because fatty acids are precursors for high-energy yielding compounds such as fatty acid methyl esters (FAMEs)2. As such, a structural and functional characterization of the protein-protein interactions of the enzymes involved in fatty acid biosynthesis serve as models for novel antimicrobials development and biofuels engineering.
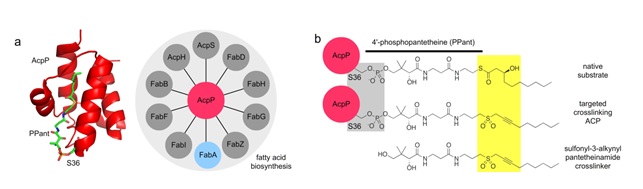
Figure 1. E. coli AcpP and crosslinking strategy. a, AcpP is a small, acidic protein comprised of four a-helices that interacts with at least 19 catalytic enzymes, 12 of which belong to FAS (10 shown here). The apolar interior of helix II and III form a hydrophobic cavity that sequesters the growing metabolite attached to the PPant arm. b, (top) A native substrate of FabA and (middle) modified AcpP with targeted sulfonyl-3-alkynyl crosslinking probe, derived from (bottom) the crosslinking pantetheinamide analog.
The synthesis of fatty acids in bacteria requires eight core fatty acid biosynthesis (Fab) enzymes and an acyl-carrier protein (AcpP) (Figure 1a). The Fab enzymes behave in dynamic but regulated manners during biosynthesis, with AcpP playing central roles3. AcpP stabilizes and shuttles the intermediate fatty acid chain to each of the eight domains that catalyze elongation and tailoring steps on the growing fatty acid chain (Figure 1a)4. These steps are determined by AcpP-Fab enzyme interactions during which AcpP chooses the appropriate catalytic partners. However, the transient nature of AcpP-Fab enzyme interactions represents major obstacles to gaining high-resolution structural and functional information about the regulatory processes involved in fatty acid biosynthesis and little is known about the AcpP-partner enzyme selection process. A new strategy is required to properly study protein-protein interactions involved in fatty acid biosynthesis.
In this work, we describe the application of a mechanism-based probe that allows active site-selective covalent crosslinking of AcpP to FabA, the E. coli acyl carrier protein (ACP) and fatty acid 3-hydroxyacyl-ACP dehydratase. This allows for covalent tethering of the AcpP and FabA active sites and capturing the AcpP=FabA complex in biologically relevant conformations. We report the 1.9 Å crystal structure of the crosslinked AcpP=FabA complex as a homodimer along the FabA dimer interface, in which AcpP exhibits two different conformations representing two snapshots of its actions (Figure 2).
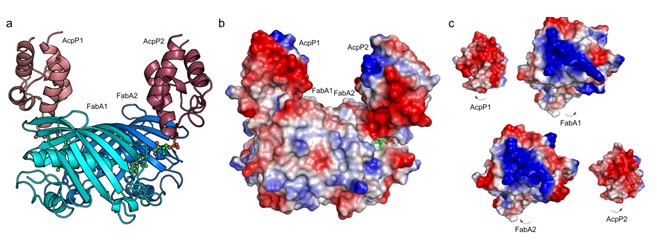
Figure 2. Structure of crosslinked AcpP=FabA. a, X-ray crystal structure of AcpP=FabA at 1.9 Å. b, The molecular surface mapped with calculated vacuum electrostatic potential of AcpP=FabA. Blue shading indicates electro-positive and red shading indicates electro-negative protein surfaces. c, Electrostatic views of protein-protein interfaces between the two pairs of AcpP=FabA dimers showing the surface differences between them.
Nuclear magnetic resonance (NMR) titration studies and accelerated molecular dynamics (AMD) (data not shown) not only support our interpretation of crystal structures but also provide an animated view of AcpP in action during fatty acid dehydration. Taken together, the crystal structure, NMR and AMD allowed for the first characterization of a conserved AcpP recognition surface and the determination of the transition between AcpP substrate sequestration and presentation, which we coined “the switch blade model”.
The structural differences observed in the enzyme complex show a stable and a destabilized interface between AcpP and FabA (Figure 2). These structural differences illustrate two active conformations between AcpP and FabA in which AcpP2 is making full contact with FabA2 while AcpP1 is in transition with FabA1. The stable interface represents the interaction between AcpP and FabA during FabA-catalyzed fatty acid dehydration while the destabilized surface represents AcpP leaving FabA’s active site. Since AcpP and FabA are covalently attached, the tethered AcpP1 is destabilized, allowing for structural determination of two active AcpP-FabA conformations.
Extensive sequence conservation among carrier proteins suggest that the mechanistic insights gleaned from these studies will prove general for fatty acid, polyketide and non-ribosomal biosyntheses. Here, the foundation is laid for defining the dynamic action of carrier-protein activity in primary and secondary metabolism, providing insight into pathways that can play major roles in biofuel and pharmaceutical developments.
The data for the final structural determination of the AcpP-FabA structure were collected at SSRL’s Beam Line 12-2. The AcpP-FabA crystals grew to 200 microns in two dimensions and less than 10 microns in the third dimension. The flatness of the crystals was a major obstacle in obtaining complete diffraction data for data processing. However, the high flux provided by Beam Line 12-2 enabled complete data collection, processing and structural determination of the AcpP-FabA complex, the first high-resolution structure of an AcpP-fatty acid enzyme interaction.
- M. Arita, "The Metabolic World of Escherichia coli is not Small", Proc. Natl. Acad. Sci. USA. 101, 1543 (2004)
- P. Nawabi, S. Bauer, N. Kyrpides, and A. Lykidis, "Engineering Escherichia coli for Biodiesel Production Utilizing a Bacterial Fatty Acid Methyltransferase", Appl. Environ. Microbiol. 77, 8052 (2011)
- K. Magnuson, S. Jackowski, C. O. Rock, and J. E. Cronan Jr., "Regulation of Fatty Acid Biosynthesis in Escherichia coli", Microbiol. Rev. 57, 522 (1993)
- D. I. Chan, and H. J. Vogel, "Current Understanding of Fatty Acid Biosynthesis and the Acyl Carrier Protein", Biochem. J. 430, 1 (2010)
C. Nguyen, R. W. Haushalter, D. J. Lee, P. R. L. Markwick, J. Bruegger, G. Caldara-Festin, K. Finzel, D. R. Jackson, F. Ishikawa, B. O’Dowd, J. A. McCammon, S. J. Opella, S.-C. Tsai, and M. D. Burkart, “Trapping the Dynamic Acyl Carrier Protein in Fatty Acid Biosynthesis”, Nature 505, 427 (2014), DOI: 10.1038/nature12810.

