An enzyme is an efficient catalyst capable of accelerating a chemical reaction in an aqueous environment under mild conditions. It not only plays a vital role in sustaining life, but also provides a tool for living organisms to create complex chemical compounds, such as antibiotics and pheromones, that can improve the biological fitness of those organisms.
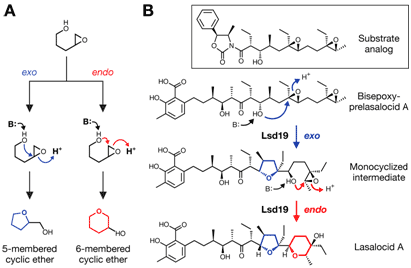
In most cases, enzymes simply accelerate the natural course of chemical reactions and minimize the occurrence of unwanted side reactions to keep the system—i.e., life—running efficiently. In rare cases, however, enzymes appear to have evolved to steer the course of chemical reactions away from the naturally favored direction to generate products that cannot be obtained otherwise. One such example is found in the biosynthesis of a class of compounds called ionophore polyether natural products. These compounds are exemplified by the presence of cyclic ethers in their structure. It was originally proposed by Westley et al. in 1974 (1) that those cyclic ethers are formed by a chemical reaction called epoxide-opening cyclization. While an epoxide-opening cyclization can proceed through two different pathways (Fig. 1a), according to Baldwin's rules of ring closure, which is a set of empirical rules proposed by Sir Jack Baldwin in 1976 that determines the feasibility of a given ring formation (2), formation of the larger ring product (endo-type cyclization, shown in red) is strongly disfavored such that only the formation of a smaller ring product (exo-type cyclization, shown in blue) is observed. Indeed, the vast majority of cyclic ethers found in ionophore polyethers are the favored exo product. However, a few occurrences of endo-type cyclic ethers are also observed.
In 1951, an ionophore polyether antibiotic named lasalocid A was isolated from a soil-dwelling bacterium Streptomyces lasaliensis (3). This compound contains both 5-exo and 6-endo cyclic ethers(Fig. 1b). In 2008, Hideaki Oikawa and his colleagues from Japan identified that an epoxide hydrolase Lsd19 catalyzes both exo- and endo-cyclization reactions to convert the bisepoxide precursor into lasalocid A (4). In order to gain a detailed understanding of how Lsd19 catalyzes the disfavored 6-membered ring formation, we set out to determine the x-ray structure of this enzyme.
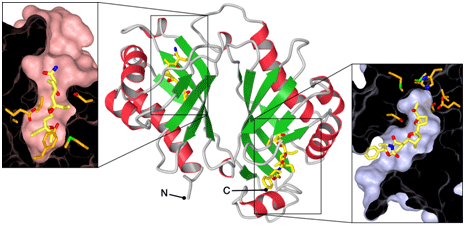
Using SSRL Beam Line 7-1 with extensive help from SSRL Staff Scientist Irimpan Mathews, we were able to determine the structure of Lsd19 in complex with a substrate and a product analog to 1.59 Å resolution. The structure revealed that the enzyme is comprised of two homologous domains belonging to the nuclear transport factor 2-like superfamily (5), with the N- and C-terminal domain catalyzing the 5- and 6-membered ring formation, respectively (Fig. 2). The structure also revealed that those highly homologous domains achieve fine substrate discrimination by adjusting the depth of their active site pocket so that only the correct substrate can bind in the corresponding active site for the intended cyclization to take place (Fig. 2, left vs. right panel). However, both reactions appear to be catalyzed by the same, simple mechanism of general acid–base catalysis. In both cases, an aspartate residue acts as a base that activates the hydroxyl group on the substrate for a nucleophilic attack on the epoxide carbon, while two other residues act as acids that stabilize the negative charge that develops on the epoxide oxygen during the transition state of the reaction.
To better understand how Lsd19 achieves catalysis of the disfavored endo-cyclization, Kendall Houk and Robert Paton from UCLA examined the system by quantum mechanical calculations. This study identified that the precise positioning of the catalytic groups within the active site is what allows the enzyme to lower the activation energy for the endo-cyclization pathway over the favored exo pathway to accomplish the predominant formation of the 6-membered ring product in the lasalocid A biosynthesis.
By combining structural and computational studies, we were able to shed light on how an enzyme can reverse the course of a chemical reaction to generate interesting and useful compounds with potent biological activities. The knowledge gained here will be valuable not only in gaining more complete understanding of the polyether natural product biosynthesis, but also in engineering enzymes that can efficiently catalyze reactions that are difficult to carry out using conventional synthetic techniques.
1. J. Berger, A.l. Rachlin, W.E. Scott, L.H. Sternbac & M.W. Goldberg. The Isolation of Three New Crystalline Antibiotics from Streptomyces. J. Am. Chem. Soc. 73, 5295–5298 (1951).
2. J.E. Baldwin. Rules for Ring Closure. J. Chem. Soc. Chem. Comm. 734–736 (1976).
3. J.W. Westley, J.F. Blount, R.H. Evans, Jr., A. Stempel & J. Berger. Biosynthesis of lasalocid. II X-ray Analysis of a Naturally Occurring Isomer of lasalocid A. J. Antibiot. 8, 597–604 (1974).
4. Y. Shichijo, et al. Epoxide Hydrolase Lsd19 for Polyether Formation in the Biosynthesis of lasalocid A: Direct Experimental Evidence on Polyene Polyepoxide Hypothesis in Polyether Biosynthesis.J. Am. Chem. Soc. 130, 12230–12231 (2008).
5. Marchler-Bauer, A. et al. CDD: Specific Functional Annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 (2009).
Kinya Hotta, Xi Chen, Robert S. Paton, Atsushi Minami, Hao Li, Kunchithapadam Swaminathan, Irimpan I. Mathews, Kenji Watanabe, Hideaki Oikawa, Kendall N. Houk & Chu-Young Kim "Enzymatic Catalysis of Anti-Baldwin Ring Closure in Polyether Biosynthesis" Nature (2012) doi:10.1038/nature10865

