The molecular structure of proteins and their complexes with other molecules are intimately linked to their biological function. Biomedical scientists and biochemists seek the structures of biological macromolecules in order to enhance their understanding of their mechanism of action at the molecular level. Structural knowledge serves many purposes, including the design of new drugs that selectively target specific biochemical reactions; the engineering of faster enzymes for industrial applications; an improved understanding of how and why abnormally-shaped proteins are involved in disorders; and the investigation of the role of certain viral proteins during infection.
At SSRL, the Joint Center for Structural Genomics (JCSG)—part of the Structural Genomics Division—and the Structural Molecular Biology Division are developing new technologies to speed up the structure determination of biological macromolecules. When x-ray beams generated by a high-energy synchrotron strike protein crystals, the x-rays are diffracted from the periodic arrangement of protein molecules in the crystal. The positions and intensities of these diffracted x-rays, measured with state-of-the-art detectors, allow researchers to compute a three-dimensional electron density map of the protein molecule, which in turn allows for the mapping of individual atoms. The quality of the electron density map ultimately determines the accuracy of the atomic positions, and hence the overall quality of the protein structure.
One approach used to calculate the electron density map from the diffracted intensities is known as molecular replacement (MR). In the MR procedure, a previously determined structure that is believed to be similar to the unknown structure is used as a reference for electron density map calculation. However, MR has its limitations—for example, traditional MR approaches tend to break down unless the unknown structure has a very close structural homolog as reference. However, data collected with the JCSG at SSRL has been used recently to develop new methods that circumvent these limitations.
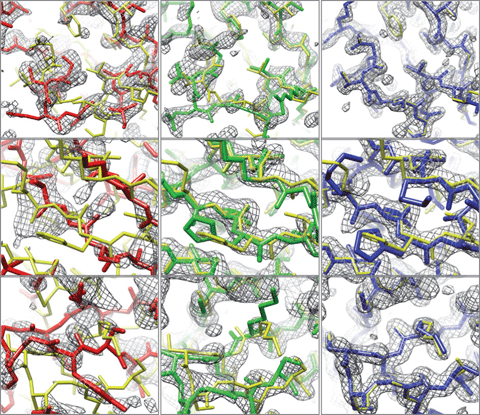
The MR approach often fails for more divergent reference models, particularly where the amino acid sequence identity between the MR reference model and the target protein is <30%. Even if MR does work in these cases, the electron density maps may be poor or they may be biased and closely resemble the structure of the reference model (see Figure). Automated tracing of an atomic model from these poor maps may result in an inaccurate model for the protein target under investigation. To help reduce mistakes in the automated tracing and enhance the capability to use more divergent reference models for MR, an international team of researchers led by Drs. Frank DiMaio and David Baker at the University of Washington in Seattle, and Thomas Terwilliger at the Bioscience Division of Los Alamos National Laboratory have combined computer algorithms for protein structure modeling with those for macromolecular structure determination by x-ray crystallography. The development and implementation of this new procedure is described recently in a letter to the journal Nature.1
Professor Baker and his co-workers reasoned that even poor electron density maps contain considerable information in those portions of the starting model that are outside of electron density and are most likely incorrect. Several years ago, Baker developed a structure prediction algorithm called Rosetta. Based on physically realistic force fields, Rosetta searches for the lowest energy conformations possible of a polypeptide chain from folding, twisting and packing the chain into a three-dimensional (3-D) molecule. Baker and colleagues proposed that Rosetta-based structure predictions guided even by poor electron-density maps from MR solutions could be used to improve an MR model. In this new procedure, energy-optimized Rosetta models were compared for consistency with the x-ray diffraction intensities, which led to improved electron density maps. The new maps were then subjected to automatic chain tracing, which produced more accurate three-dimensional atomic models of the protein structure. These models were then handed back to JCSG crystallographers to complete the structure determination.
This new Rosetta-based method was tested on eight sets of x-ray diffraction data, the structures for which had previously been elusive. Three of the eight datasets used in this study were acquired at SSRL as part of JCSG’s program under the NIGMS Protein Structure Initiative. The Figure demonstrates the improvement in the electron density map and structural accuracy before and after the application of the Rosetta-guided method. This approach to MR of combining electron density map interpretation with energy minimization is likely to be increasingly useful as more x-ray diffraction datasets on proteins with more remote amino acid identity are collected. Implementation of the Rosetta-based method enhances the possibility that existing structures can successfully be used as MR models to determine the structures of these more sequence-remote proteins. This new method is now available to the public, and is distributed with the PHENIXcrystallographic software package.
1. F. DiMaio, T.C. Terwilliger, R.J. Read, A. Wlodawer, G. Oberdorfer, U. Wagner, E. Valkov, A. Alon, D. Fass, H.L. Axelrod, D. Das, S.M. Vorobiev, H. Iwai, P.R. Pokkuluri, D. Baker Improved Molecular Replacement by Density- and Energy-guided Protein Structure Optimization Nature 473, 540–543 (2011)
2. P.D. Adams, P.V. Afonine, G. Bunkóczi, V.B. Chen, I.W. Davis, N. Echols, J.J. Headd, L.W. Hung, G.J. Kapral, R.W. Grosse-Kunstleve, A.J. McCoy, N.W. Moriarty, R. Oeffner, R.J. Read, D.C. Richardson, J.S. Richardson, T.C. Terwilliger, P.H. Zwart PHENIX: A Comprehensive Python-based System for Macromolecular Structure Solution. Acta Cryst. D66, 213-221 (2010).
F. DiMaio, T.C. Terwilliger, R.J. Read, A. Wlodawer, G. Oberdorfer, U. Wagner, E. Valkov, A. Alon, D. Fass, H.L. Axelrod, D. Das, S.M. Vorobiev, H. Iwai, P.R. Pokkuluri, D. Baker Improved Molecular Replacement by Density- and Energy-guided Protein Structure Optimization Nature 473, 540–543 (2011)

