Genetic information is read by RNA polymerase II (Pol II) by synthesizing messenger RNA from DNA in the process termed transcription. Transcription is triggered by the binding of transcription factor(s) to specific DNA regulatory sequences, resulting in a recruitment of Pol II to a specific gene to initiate transcription. Since transcription by Pol II accounts for biological activities in eukaryotes, elucidation of the transcription process is essential to understanding biology as a whole. Mediator is a large multi-protein complex that regulates most, if not all, gene transcription by Pol II.1 It connects activator and repressors bound as regulatory DNA sequences to Pol II and a set of general transcription factors (GTFs) that assemble at the promoter for transcription initiation1, 2. Mediator is structurally and functionally conserved in all eukaryotes3. In the yeast S. cerevisiae, where Mediator was first discovered, Mediator comprises 21 subunits with a total mass over 1 mega Daltons4, and is organized in three distinct modules, termed Head, Middle/Arm, and Tail5
One of the outstanding questions regarding the mechanisms of Mediator functions is how 21 subunits assemble together to form a large multi-protein complex stably, and how they together bring about transcription regulation. Over the years, the size and complexity of Mediator has hampered our understanding of Mediator assembly and its role in regulating transcription.
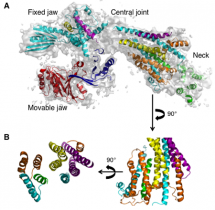
By using state-of-the-art protein complex engineering and by collecting diffraction data at the Stanford Synchrotron Radiation Lightsource and Advanced Photon Source, the crystal structure of an essential Mediator sub-complex—Head module (7 subunits with molecular mass 223 kDa) was determined at 4.3 Å resolution6 (Figure 1). The Head structure shows three distinct domains, termed Fixed Jaw, Movable Jaw, and Neck, that come together to form a flexible central joint region (Figure 1A).
This structure has provided the answer to the key question of how each subunit assembles to form a stable complex. Strikingly, a portion of the Head named the Neck domain provides the stability and integrity of the complex by arranging the five proteins in a polymer-like structure, termed alpha-helical bundles (Figure 1B). An intricate pattern of interactions within this helical bundle ensures the stable assembly of the Head subunits, and provides the binding sites for general transcription factors (GTFs) and Pol II—a unique assembly mechanism, possibly transforming a common view multi-protein complexes—from the simple sum of each protein to a highly integrated assembly of many. The results, in essence, reveal architectural principles underlying the role of Mediator in the regulation of gene expression. The results will have broad significance, inasmuch as all the Mediator proteins involved are conserved from yeast to humans. Moreover, the ability to solve such complex structures is important because multi-protein complexes such as Mediator will most likely become next generation of drug targets.
As the structures of Pol II and ribosomes have already been solved, a high-resolution structure determination of Mediator (21 subunits, 1 mega Da) is one of the remaining grand challenges to be achieved in structural biology. This work represents a major step toward the future structure determination of the entire Mediator.
This research was supported by NSF grant MCB 0843026 and the AHA 0735395N to Y.T. T.I. was supported by Human Frontier Science Program long-term fellowship. NIH grants R01 GM67167 to F.J.A., GM36659 to R.D.K. NCI Cancer Center Support Grant P30 CA08748 to MSKCC Microchemistry and Proteomics Core Laboratory, and the European Commission Framework Program 7 projects INSTRUCT and P-CUBE to I.B. Crystallography data was collected at the Stanford Synchrotron Radiation Lightsource and the Advanced Photon Source, supported by DOE, OBER, NIH, NCRR, Biomedical Technology Program, and the NIGMS.
1. R. D. Kornberg, Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30, 235 (May, 2005).
2. S. Malik, R. G. Roeder, The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11, 761 (Nov, 2010).
3. M. Boube, L. Joulia, D. L. Cribbs, H. M. Bourbon, Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110, 143 (Jul 26, 2002).
4. S. Bjorklund, C. M. Gustafsson, The yeast Mediator complex and its regulation. Trends Biochem Sci 30, 240 (May, 2005).
5. G. Cai et al., Mediator head module structure and functional interactions. Nature structural & molecular biology 17, 273 (Mar, 2010).
6. T. Imasaki et al., Architecture of the Mediator head module. Nature 475, 240 (Jul 14, 2011).
T. Imasaki, G. Calero, G. Cai, K.-L. Tsai, K. Yamada, F. Cardelli, H. Erdument-Bromage, P. Tempst, I. Berger, G. Lorch. Kornberg, F. J. Asturias, R. D. Kornberg and Y. Takagi, "Architecture of the Mediator Head Module", Nature 475, 240 (2011)




