
How does a protein distinguish its target substrate from other substrates? For flap endonuclease (FEN), it must distinguish a 5′ single-stranded (ss)DNA or RNA flap, a structure formed during Okazaki fragment maturation in lagging strand replication, from intact dsDNA, ssDNA, RNA, 3′ flaps, DNA bubbles, and DNA ends, while allowing either RNA or DNA be in the 5′ flap (Fig. 1). It also incises one nucleotide into the double stranded (ds) region, creating a ligatable product for efficient resolution of the flap. FEN1 has primary responsibility for removing the estimated 50 million short DNA flaps created during replication of mammalian nuclear genomes. It is part of a nuclease superfamily that recognizes ss-ds junctions in nucleic acid substrates and they all incise one nucleotide into the ds region. Xeroderma pigmentosum complementation group G protein (XPG), Exonuclease 1 (EXO1), and Gap endonuclease (GEN) are superfamily members that recognize DNA bubbles, ends of DNA, and Holliday junctions, respectively.1,2
Much of the focus on substrate recognition has been on the single strand of the 5′ flap, with the mechanism based on initial ss binding, ds incision. However, recent crystallographic determination of the protein:DNA complex revealed a different mechanism—ds binding, ss incision.3 John Tainer’s group at the Scripps Research Institute in La Jolla and at Lawrence Berkeley National Laboratory screened multiple crystals and collected data at SSRL Beam Line 11-1 and ALS Beam Line 12.3.1, determining the first product and substrate DNA complexes. The structures provided the first complete picture of how FEN binds to DNA (Fig. 2A). The active site held two metals, coordinated directly and indirectly by seven carboxylates, all highly conserved in the FEN1 superfamily. The DNA is bound across the whole protein and is remarkably bent 100° at the junction break (Fig. 2B). This dramatic bend over one nucleotide selects against intact dsDNA. The positioning of the bend positions the 5′ flap at the active site. An unpaired 3′ flap is held in a pocket that binds primarily to the backbone and with no base specificity. The unpaired 3′ flap is critical for creating a product that can be ligatable, since incision occurs one nucleotide into the dsDNA region.
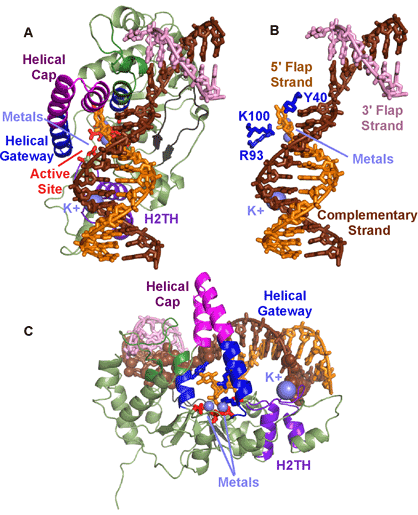
The major interaction is not to the strand that is incised but to the strand complementary to it (Fig. 2C). The bias of the binding to the non-incised strand selects for duplex DNA, avoiding inadvertent incision of ssDNA that may have resulted if primary interaction was to the incised strand. This relative lack of interaction to the incised strand allows either RNA or DNA to be within the incised strand. Thus we propose that binding is through dsDNA interactions, albeit to dsDNA with a break that can bend to such a dramatic angle. Additionally, the major dsDNA contact points are not contiguous but are separated by a distance of about 23-24 Å. On one side is a K+ coordinated by a conserved H2TH motif. On the other side is the bend. This separation of binding sites not only provides another validation for dsDNA, it selects for 5′ flaps. For a 3′ flap to be positioned at the active site, the separation between dsDNA contact points must be more narrow.
Fen selectivity for ss flaps with free termini are encoded in the helical gateway that guards the active site (Fig. 2C). Its narrow width would only allow ssDNA or RNA to access the active site. A helical cap covers the gateway, thus selecting for free termini that can thread through the gateway under the cap. Interestingly, the gateway is conserved in the FEN superfamily—which all incise substrates with single stranded regions. The cap is FEN1 and EXO1 specific, consistent with the free terminal end in the substrates for those two superfamily members
Three protein:DNA complex structures were determined that revealed an unexpected mechanism for incision. The two substrate structures showed that the DNA on either side of the scissile phosphate were paired and the scissile phosphate itself was distant from the catalytic metals while the product structure showed that the terminal base of the product was unpaired. For the scissile phosphate to be near the catalytic metals, two nucleotides must be unpaired. Thus, we propose that incision actually occurs in ss nucleotides.
The structure-derived mechanism is consistent with biochemical data and helped to explain certain inconsistencies in the biochemistry. Further work is clearly needed to prove and provide more details on the mechanism, but the structural analysis has provided invaluable information on the selectivity of the flap endonuclease family.
Work on FEN1 is supported by the NIH/NCI through RO1CA081967, R01CA073764, and P01 CA092584 (Structural Biology of DNA Repair Program Project) and by BBSRC (BBF0147321). Crystal data was collected at the SIBYLS Beam Line 12.3.1 supported by the IDAT program (ALS, Contract DE-AC02-05CH11231) and Beam Line 11-1 (SSRL, supported by DOE, OBER, NIH, NCRR, Biomedical Technology Program, and the NIGMS).
1- Lieber, 1997 M.R. Lieber, The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19 (1997), pp. 233–240
2 - Tomlinson et al., 2010 C.G. Tomlinson, J.M. Atack, B.R. Chapados, J.A. Tainer and J.A. Grasby, Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem. Soc. Trans., 38 (2010), pp. 433–437.
3 - Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. (2011) Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 145,198-211.
Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. (2011) Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 145,198-211.




