For more than 60 years, doctors have used compounds produced by the penicillium mold as a means of fighting off harmful bacteria, and for almost the same length of time we have employed molecules produced by soil bacteria in our clinics as antibiotics (aminoglycosides and macrolides) to treat infection. Yet bacterial defense mechanisms, including a property unique to bacteria known as horizontal gene transfer, fight back and resistance to these chemicals has become commonplace in hospitals, rest homes, and the general population.
In the early 1940s, when penicillin first became available for clinical use, less than 1% of Staphylococcus aureus strains showed resistance—not surprising in that the bacteria had never encountered penicillin and had no defense mechanism against it. This rose to 14% by 1946 and 38% a year later. Today more than 90% of all S. aureus infections are resistant to penicillin. Methicillin was developed to control S. aureus but now methicillin-resistant strains (MRSA) are also commonplace around the world. MRSA is not only resistant to methicillin but to most penicillin-like drugs (the so-called β-lactam antibiotics) and even to some of the other clinically relevant antibiotics including erythromycin and vancomycin. MRSA is the classic example of what we call a “super-bug.” Subsequent drug discovery has led to the development of four classes of β-lactam antibiotics including the carbapenems, classified as “last line of defense” drugs for E. coli and Klebsiella pneumoniae infections. Since there are no new antibiotics in the pipeline to combat bacteria resistant to carbapenems, the worldwide spread of resistance to these compounds is considered a potential nightmare scenario.
Resistance to the β-lactam antibiotics is primarily due to the production of enzymes which destroy the active part of the drug. These enzymes, known as β-lactamases, have evolved from a family of enzymes (called PBPs) that the bacteria use to build and maintain their strong cell wall structures. These PBP enzymes are the biological target of penicillin drugs, where the drug attaches to the enzyme, making it unable to function and ultimately weakening the cell wall until the bacteria ruptures and dies. Mutations in the PBPs have produced the β-lactamase enzymes which will preferentially bind the penicillin drugs and break them apart, before the drugs can get a chance to attach to the PBPs. Although it seems reasonable to speculate that evolution of β-lactamase enzymes is predominantly the result of the use of penicillin drugs, we have discovered that a previously unknown bacterial strain (Oceanobacillus iheyensis) found in deep ocean sediment (a place where it is highly unlikely that the bacteria would have ever encountered man-made penicillins) has a very active β-lactamase enzyme (designated Oih-1). We solved the structure by molecular replacement using diffraction data to 1.25 Å resolution collected at SSRL Beam Line 12-21 and showed that Oih-1 has a three-dimensional structure almost identical to the known β-lactamases in common bacteria, although it also shows adaptive features representative of a life spent at the bottom of the ocean, namely an extremely high tolerance to salt and a highly negatively charged molecular surface. Clearly the development of Oih-1 cannot be due to exposure to clinically-derived β-lactam antibiotics so it is likely that evolution from the PBPs to the β-lactamases took place prior to the intervention of man, and as a result of the ongoing struggle for survival in the microscopic world.
Most bacterial strains harboring these β-lactamase enzymes were resistant to all the older β-lactam antibiotics but seemed to be inactive against the “last resort” carbapenems, but in recent years the occurrence of carbapenem-resistant strains of bacteria has been reported. The underlying molecular cause is predominantly β-lactamase enzymes that have developed the ability to break down the carbapenems, including the K. pneumoniae carbapenemase (KPC), the New Delhi metallo-β-lactamase (NDM-1), and the Guiana extended-spectrum β-lactamases (GES) also from K. pneumoniae. The NDM-1 enzyme is extremely worrisome in that it is capable of breaking down all known classes of β-lactam antibiotics, which necessitates the use of other types of antibiotics. Unfortunately the bacteria from which NDM-1 has been isolated contain other resistance elements making them also resistant to most other drugs including the aminoglycosides and the fluoroquinolones, making these bugs, like MRSA, true “superbugs.”
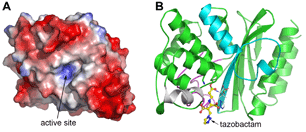
The GES family consists of 15 known variants, all point mutations of the first identified member, GES-1, the structure of which we reported in 20072 using data collected at SSRL Beam Lines 7-1 and 11-1. Although the GES-1 three-dimensional structure is similar to other β-lactamase structures which have been reported, it contains a highly specific disulfide bridge next to the site of enzyme activity which has been linked in KPC to carbapenemase activity. Although GES-1 was unable to break down imipenem (the simplest of the carbapenem drugs), subsequent members of the family (GES-2, GES-4, GES-5, GES-6 and GES-14) all have increasing carbapenemase activity, with the latter enzyme having the dubious distinction of being capable of breaking down all known β-lactams, and putting it on par with NDM-1. Fortunately the GES enzymes can be stopped in their tracks by known β-lactamase inhibitors and we have recently demonstrated this with the high resolution structure of GES-2 with tazobactam, using data collected at SSRL Beam Line 12-2. The tazobactam forms a tight covalent complex with GES-2 and there is some evidence that the inhibitor breaks down and sticks in a nearby part of the enzyme, effectively preventing the attachment of other β-lactam drugs.
It appears that the GES family of enzymes is evolving the ability to break down carbapenems which, while alarming, is not unexpected given the “pressure” that we are applying to the environment with respect to the use, overuse and misuse of antibiotics which will naturally result in mutation, adaption and evolution of resistance mechanisms. The bacteria housing the genes for the GES enzymes can undergo cell division thousands of times a year, giving ample opportunity for evolution to take hold and produce highly active mutant strains. This, along with horizontal gene transfer commonplace in nature and rampant in hospitals and clinics, will continue to give rise to “superbugs” that do not respond to treatment and require the continued development of new drugs.
Toth, M., Smith, C., Frase, H., Mobashery, S., Vakulenko, S. (2010). An Antibiotic Resistance Enzyme from a Deep-Sea Bacterium. J. Am. Chem. Soc.132, 816–823.
Smith, C.A., Caccamo, M., Kantardjieff, K.A., Vakulenko, S. (2007). The Crystal Structure of GES-1 at Atomic Resolution: Insights into the Evolution of Carbapenamase Activity in the Class A Extended-Spectrum b-Lactamases. Acta Crystallogr. D63, 982-992.
Frase, H., Smith, C.A., Toth, M., Champion, M.M., Mobashery, S., Vakulenko, S.B. (2011). Identification of Products of Inhibition of GES-2 β-Lactamase by Tazobactam by X-Ray Crystallography and Spectrometry. J. Biol. Chem. 286, 14396-14409.

