Despite having access to only a handful of metal coordinating groups, the ability of proteins to control and harness the reactivity of metal centers is unmatched in small molecule metal complexes. This is primarily due to the fact that the coordination and solvation environments of metals can be exquisitely tuned by the highly evolved non-covalent bonding networks of proteins that surround the metal sites.
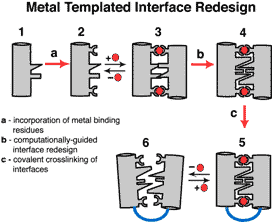
With this in mind, we developed a rational engineering approach, Metal Templated Interface Redesign (MeTIR). MeTIR is based on the initial complexation of non-self-interacting proteins by metal coordination, followed by the re-engineering of the metal-templated interfaces for stability and selectivity (Figure 1). Using the protein design software Rosetta, we incorporated favorable non-covalent interactions into the interfaces of Zn4:MBPC-14. The resulting assemblies (Zn4:RIDC-14, PDB ID: 3HNI and Zn4:RIDC-24, PDB ID: 3HNJ) display significant stabilization over the parent complex while retaining the original supramolecular architecture, indicating that our “template-and-stabilize” approach works as planned. Significantly, we found that the non-covalent interactions built into the interfaces are favorable enough to hold the RIDC-1 and RIDC-2 building blocks together even in the absence of metal coordination (PDB ID: 3HNK) with micromolar dissociation constants, indicating that metal-templating may provide a straightforward path to designing de novo protein interfaces, a currently unsolved problem in biochemistry. With this in mind, we developed a rational engineering approach, Metal Templated Interface Redesign (MeTIR). MeTIR is based on the initial complexation of non-self-interacting proteins by metal coordination, followed by the re-engineering of the metal-templated interfaces for stability and selectivity (Figure 1). Using the protein design software Rosetta, we incorporated favorable non-covalent interactions into the interfaces of Zn4:MBPC-14. The resulting assemblies (Zn4:RIDC-14, PDB ID: 3HNI and Zn4:RIDC-24, PDB ID: 3HNJ) display significant stabilization over the parent complex while retaining the original supramolecular architecture, indicating that our “template-and-stabilize” approach works as planned. Significantly, we found that the non-covalent interactions built into the interfaces are favorable enough to hold the RIDC-1 and RIDC-2 building blocks together even in the absence of metal coordination (PDB ID: 3HNK) with micromolar dissociation constants, indicating that metal-templating may provide a straightforward path to designing de novo protein interfaces, a currently unsolved problem in biochemistry.
Ultimately, MeTIR leads to the formation of a stable framework around a metal. An implication of this metal-templated supramolecular rigidification is an increase in the affinity and specificity for the templating metal ion by the superprotein scaffold. To probe whether or not this is indeed the case, we created the next-generation variant of RIDC-1,C96RIDC-1, which was designed to form a Zn-templated assembly stabilized with a combination of covalent and non-covalent interactions across its interfaces.1 C96RIDC-1 was found to form a tetramer (C96RIDC-14, Figure 2a, PDB ID: 3IQ5) even in the absence of Zn coordination with a dissociation constant of 50 nM. This makes C96RIDC-14 one of the most stable protein complexes ever engineered. Upon Zn-binding, C96RIDC-14 undergoes a large structural change reminiscent of natural switch proteins (Figure 2a, PDB ID: 3IQ6). Importantly, C96RIDC-14 displays nanomolar Zn affinity (which represents over a 1000-fold increase compared to the parent MBPC-1), and exclusive Zn selectivity over other divalent metal ions including Co(II), Ni(II) and Cu(II) (Figure 2b and Figure 3). The selectivity of Cu(II) is particularly noteworthy, as there is no synthetic ligand system–to our knowledge–that can exclusively bind Zn(II) over Cu(II).
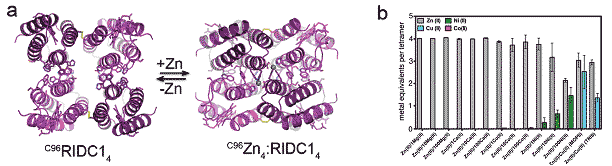
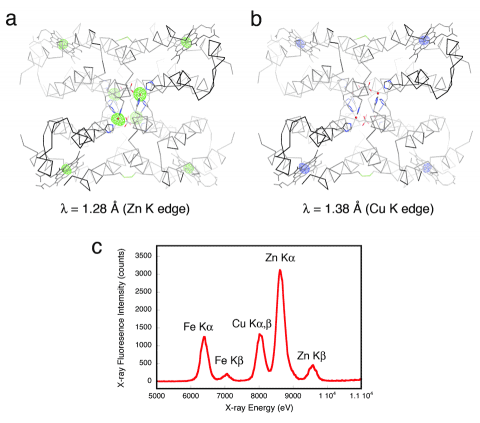
A converse implication of metal-templated supramolecular rigidification is that metal ions with alternative stereochemical preferences should be disfavored and forced into high-energy conformations, which could lead to reactivity. Accordingly, we have found that Cu(II) ions are forced into an unsaturated coordination geometry with the RIDC-1 construct (PDB ID: 3HNL), which was designed based on Zn-templating.
Our combined findings with MeTIR thus highlight another important advantage of using proteins as ligand platforms, namely the possibility of employing their extensive, chemically rich surfaces to tune the coordination chemistry and reactivity of interfacial metal ions. Simultaneously, the observation that MeTIR can lead to such structural and diversity with minimal mutations on a protein surface raises the question whether similar templating steps may have been operative in the natural evolution of proteins (metallo- or non-metallo) and protein complexes.
All of the aforementioned crystal structures, which form the basis of all of our conclusions and validate our theories, were determined using SSRL beamlines. These beamlines (9-2 and 7-1) not only ensured high resolution structures to be obtained, but their multi-wavelength and X-ray fluorescence detection capabilities enabled an unambiguous characterization of the metal centers formed in our protein complexes.
1. Brodin, J. D.; Medina-Morales, A.; Ni, T.; Salgado, E. N.; Ambroggio, X. I.; Tezcan, F. A., J. Am. Chem. Soc. 2010, 132, 8610-8617.
Salgado, E. N.; Ambroggio, X. I.; Brodin, J. D.; Lewis, R. A.; Kuhlman, B.; Tezcan, F. A., Proc. Natl. Acad. Sci. USA 2010, 107, 1827-1832.




