Copper is a required micronutrient for all living cells, being an essential component of many metalloenzymes, but free intracellular copper is highly toxic. Because of this copper within cells is very tightly controlled, with specific copper ion pumps (both importers and exporters) located at the cell surface, which are coupled to highly specific metallochaperones that act as molecular taxi-cabs, transporting copper to the active sites of particular target proteins [1]. The expression of the copper homeostatic apparatus is also very tightly controlled, and is very sensitive to copper levels. In bacteria, two families of copper-responsive transcriptional repressors have been described; these are typified by the CueR repressor in Escherichia coli [2], and the CopY repressor in Enterococcus hirae [3]. However, many bacteria, including important human pathogens, such as Mycobacterium tuberculosis (Figure 1) and Listeria monocytogenes, contain neither family, and the mechanism of copper regulation in these organisms has been unknown. The discovery of the CsoR repressor from M. tuberculosis represents the first example of a totally new family of bacterial repressors that appears to be more widespread than either of the two established repressor families.
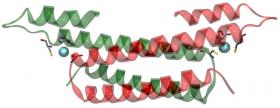
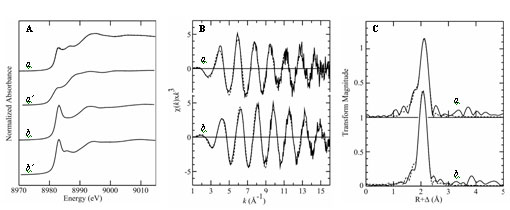
In the presence of copper the CsoR repressor dissociates from the cso operon which can then be expressed. The operon contains the gene for what is believed to be a cellular copper exporter - CtpV [4]. Binding of Cu(I) to the CsoR dimer is thought to cause a conformational change, [effecting decreased] CsoR affinity for DNA, allowing expression of the cso operon, thus providing a protective mechanism against toxic levels of copper within cells. The binding of copper to CsoR is exceptionally strong (KCu ≥ 10-19M), which is expected for a system that is critical for removal of toxic copper from cells. While CsoR binds two Cu(I) per homodimer only one copper is required to dissociate it from DNA. Liu et al used a series of techniques to understand the structure of the Cu(I) bound CsoR repressor. Protein crystallography of Cu(I)-bound CsoR indicates that CsoR is an alpha-helical dimer, with each protomer composed of three helices (Figure 2). The copper is bound between the two subunits, coordinated by the side chains of amino acids from each subunit (Cys36, Cys65˙, and His61˙). Copper K-edge X-ray absorption spectroscopy (XAS) was used to provide accurate bond-length information on the copper site of Cu(I)-bound CsoR. XAS experiments were conducted on SSRL's structural molecular biology XAS beamline 9-3, with additional (preliminary) data being measured on the Canadian Light Source HXMA beamline. Analysis of the extended X-ray absorption fine structure (EXAFS) oscillations indicated a three-coordinate site with two Cu-S ligands at 2.21 Å and one oxygen or nitrogen at 2.06 Å. Analysis of the Cu XAS of the Cu(I)-CsoR H61A mutant indicated a two-coordinate site with two sulfurs at 2.14 Å (Figure 3).
Sequence comparisons indicate that homologues of CsoR are very widespread in prokaryotes, being present in all major classes of eubacteria. The study of Liu et al thus represents a structural and functional characterization of the first known member of a new and widely-distributed family of prokaryotic copper regulators. The presence of CsoR is likely to enhance the survival of M. tuberculosis in the host, and whether CsoR can be exploited as the basis for new treatments for tuberculosis awaits future research.
- Rae, T.D., Schmidt, P.J., Pufahl, R.A., Culotta, V.C. & O'Halloran, T.V. Science 1999, 284, 805-808.
- Strausak, D. & Solioz, M. J. Biol. Chem. 1997, 272, 8932-8936.
- Stoyanov, J.V., Hobman, J.L. & Brown, N.L. Mol. Microbiol. 2001, 39,502-511.
- Palmgren, M.G. & Axelsen, K.B. Biochim. Biophys. Acta 1998, 1365, 37-45
Liu, T., Ramesh, A., Ma, Z., Ward, S. K., Zhang, L., George, G. N., Talaat, A. M., Sacchettini, G. C., Giedroc, D. P. "CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator". Nature Chem. Bio., 2007, 3, 60-68