There are over 50 mammalian cytochrome P450 genes in at least 17 families. Microsomal cytochrome P450 enzymes catalyze specific steps in the biosynthesis of steroid hormones, cholesterol, prostanoids and bile acids, participate in the catabolism of endogenous compounds, including fatty acids and steroids, and are involved in the degradation of exogenous compounds, including a wide variety of structurally diverse drugs and carcinogens. The human cytochrome P450 2A6 is principally involved in the break down of nicotine in the bloodstream as it circulates through the liver. Oxidation of nicotine by this P450 leads to detoxication, but the enzyme also activates tobacco-specific procarcinogens to mutagenic products (1-4).
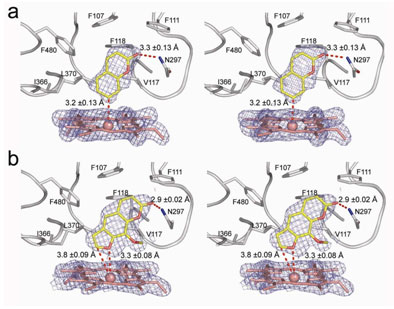
Using x-ray diffraction data sets collected on BL 11-1 and BL 9-1 at SSRL, a team of scientists at The Scripps Research Institute has solved the crystal structure of human cytochrome P450 2A6. The resources of these beam lines were employed for automated screening of many crystals, in order to identify crystals that diffracted well and at the same time had substrate or inhibitor bound to the enzyme. For the crystals that did diffract well, the parallel x-ray beams and large area detectors available on these beam lines were required to record the data, as the protein crystals not only diffracted to high resolution, but also exhibited a large unit cell. P450 2A6 structures were solved with the alternative substrate, coumarin, and with the inhibitor, methoxsalen, bound in the active site of the enzyme, adjacent to its iron containing heme group. The compact, hydrophobic active site contains one hydrogen bond donor, Asn297, which orients coumarin for regioselective oxidation (Fig. 1a). Methoxsalen also interacts with Asn297 and effectively fills the active site cavity without significantly perturbing the structure (Fig. 1b). Precise knowledge of these molecular details and interactions is critical for the rational design of new inhibitors. This structural information is being used in on-going high resolution experiments at SSRL to probe the active site of P450 2A6 with additional small molecule compounds. An effective inhibitor of P450 2A6 could be used to diminish smoking and tobacco-related cancers by reducing dependence on nicotine and by blocking formation of carcinogens.
This work was supported by National Institutes of Health Grant GM031001 (to E.F.J.) and fellowship 12FT-0185 from The Tobacco-Related Disease Research Program (J.K.Y.)
- Sellers, E.M., Kaplan, H.L., and Tyndale, R.F. Clin. Pharmacol. Ther. 68, 35-43 (2000).
- Sellers, E.M., et al. Nicotine Tob. Res. 5, 891-899 (2003).
- Patten, C.J. et al. Carcinogenesis 18, 1623-1630 (1997).
- Fujita, K. and Kamataki, T. Environ. Mol. Mutagen. 38, 339-346 (2001).
Yano, J.K., Hsu, M.-H., Griffin, K.J., Stout, C.D. and Johnson, E.F. (2005) The structure of human microsomal cytochrome P450 2A6 with coumarin and methoxsalen bound. Nature Struct. Mol. Biol. 12, 822-823.




