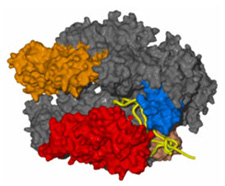
Professor Roger D. Kornberg and his group in the Stanford University School of Medicine have devoted more than 20 years to the study of the process by which genetic information encoded in all living things by DNA is processed into a message (a process called transcription that produces messenger RNA) that then directs the synthesis of proteins. A breakthrough paper detailing the structures of the core RNA Polymerase II protein was published in Science in April 2000 and followed by two more papers in Science a year later. The studies published in 2001 made use of synchrotron data from SSRL to reveal the molecular structure at a resolution of 2.7 Å and a lower resolution structure of the enzyme in the process of transcribing a fragment of DNA into RNA [1]. This pioneering work has continued and another pair of seminal papers has just appeared in Science [2,3]. A highlight summarizing the work in the same issue of Science states that "Initiation of transcription in eukaryotes involves the assembly of a large complex, composed of RNA polymerase II (pol II) and five general transcription factors, at the gene promoter. The transcription factor TFIIB plays a role in bridging between promoter DNA and pol II. Bushnell et al. (p. 983) have determined the structure of a complex of the 10-subunit yeast RNA pol II with TFIIB at 4.5 Å resolution. Using structural information available on the other four transcription factors, TFIID, TFIIE, TFIIF, and TFIIH, they propose a nearly complete model of the transcription initiation complex. Westover et al. (p. 1014) provide new insights into transcription itself, through the 3.6 Å structure of pol II in a complex with synthetic DNA and RNA oligonucleotides. The structure reveals how the RNA transcript is separated from the DNA template during transcription."
The development [4] and more recent full deployment of an automated robotic screening system at SSRL, enabled by funding provided by the National Institutes of Health, made the job of finding good diffracting crystals for these studies much faster and more efficient. This automated system uses tiny frozen crystals stored on nylon loops at the end of metal pins. A robotic arm retrieves each pin and aligns the crystal in the path of the x-ray beam. The robot can automatically test 288 samples without the need for researchers to manually transfer each sample as was done in the past. "It saves a lot of time while optimizing the quality of the data," said SSRL scientist Mike Soltis, head of the macromolecular crystallography group. "With the new system, the Kornberg group screened 130 crystals in seven hours without losing any. Two weeks earlier, they had manually mounted 100 crystals in 24 hours, losing a few crystals and much sleep in the process. The new automated screening system significantly reduces the time to complete studies of the complexity of RNA polymerase." This new automated robotic screening system will be available to all of SSRL's general users
- See http://www-ssrl.slac.stanford.edu/research/highlights_archive/rna_ polymerase.html
- D. A. Bushnell et al. Science 303, 983 (2004)
- K. D. Westover et al. Science 303, 1014 (2004)
- A. E. Cohen et al. J. Appl. Cryst. 35, 720 (2002).
D. A Bushnell, K. D Westover, R. E Davis and R. D Kornberg, "Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms", Science 303, 983 (2004)

