Botulinum neurotoxins (BoNTs) have Janus faces in that they act as both deadly toxins that cause botulism and effective medicines (e.g. Botox® and Dysport®). These toxins invade motor neurons at their junctions with muscular tissue, where they disable the release of the neurotransmitter acetylcholine and subsequently paralyze the affected muscles. This common mechanism underlies both the toxic and therapeutic functions of BoNTs. Accidental BoNT poisoning primarily occurs through ingestion of food contaminated by Clostridium botulinum, the bacterium that produces BoNTs. However, BoNTs by themselves are fragile and sensitive to low pH environments and digestive proteases. So how do they survive the harsh environment of the host’s gastrointestinal tract?
Researchers at Sanford-Burnham Research Institute and the Medical School of Hannover in Germany recently discovered that the neurotoxin has a bodyguard, a protein termed non-toxic non-hemagglutinin (NTNHA) protein, which is produced by the C. botulinum bacterium simultaneously with BoNT. This study, published in Science, revealed the first crystal structure of a ~300kDa protein complex composed of serotype A BoNT (BoNT/A) and NTNHA, which together form the minimally functional progenitor toxin complex (M-PTC) (Figure 1). NTNHA keeps the toxin safe through the gut and then releases it upon entering the bloodstream.
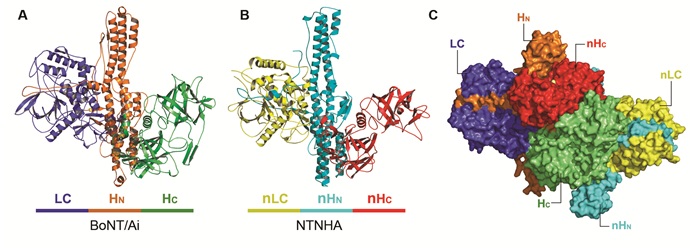
The study used a genetically modified, catalytically inactive BoNT/A that carries three mutations in its catalytic site (termed BoNT/Ai) and revealed the crystal structure of the M-PTC at 2.7 Å resolution. The method used was single isomorphous replacement with anomalous scattering (SIRAS), based on X-ray diffraction data collected at SSRL’s Beam Line 9-2 and NE-CAT at the Advanced Photon Source. Surprisingly, the non-toxic NTNHA exhibited a three-domain architecture similar to the neurotoxin, although they share less than 20% sequence identity (Figure 1). The two proteins hug each other, interlocking with what looks like a handshake and providing unusually large and multivalent binding interfaces to shield BoNT from the hostile gastrointestinal environment.
Biochemical and functional studies also revealed a novel pH-sensing mechanism by which the M-PTC, which is tightly bound at acidic pH in the gut, dissociates at neutral pH after absorption from the gut into the general circulation. Several essential residues involved in pH-dependent assembly of BoNT/A and NTNHA which were identified by structure-based mutagenesis are weak spots of the toxin that could be targeted with new therapeutics.
This discovery suggests an innovative approach for oral inhibitor design, which aims to mimick pH change, prematurely breaking up BoNT’s protective embrace and leaving the stomach’s digestive enzymes and acid to destroy the unprotected toxin. This type of therapy—either alone or in combination with other therapies currently in development—could be given preventively at a time when BoNT contamination becomes a public health concern. Furthermore, a better understanding of the toxin protection mechanism will enable the design of novel proteinaceous vehicles to allow oral administration of drugs that currently must be injected.
This work was partly supported by an Alfred P. Sloan Research Fellowship and NIH grant (R01AI091823) to RJ.
Shenyan Gu, Sophie Rumpel, Jie Zhou, Jasmin Strotmeier, Hans Bigalke, Kay Perry, Greice Krautz-Peterson, Charles B. Shoemaker, Andreas Rummel, Rongsheng Jin. (2012) "Botulinum neurotoxin is shielded by NTNHA in an interlock complex." Science, 335 (6071): 977-981.




