With the emerging proteomics era, the biosciences community is now challenged to elucidate the structures and functions of a large number of interacting proteins. Our understanding of modern biology has to a significant degree resulted from our knowledge of the structures of biological molecules, which provide direct information relating to function. The elucidation of in vitro supramolecular structures —spanning length scales from nanometers to microns— designed to model in vivo conditions of interacting proteins required for cell function, will require interdisciplinary approaches and techniques. An important goal in biophysics is the understanding of interactions leading to supramolecular structures of cytoskeletal proteins and associated biomolecules, and most important, the elucidation of the roles the structures play in cell functions.
Our group at the University of California at Santa Barbara (UCSB) has recently reported on the structure of filamentous (F) actin complexed with the actin cross-linking protein a-actinin (1). On the nanometer scale, synchrotron x-ray scattering and diffraction carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), has revealed a structural transition from a loose network of filaments at low cross-linker concentrations, to a disordered quasi-square lattice of actin fibers within bundles, at cross-linker concentrations near physiological concentrations. On the micron scale, laser scanning confocal microscopy has revealed a relatively rigid, frequently branching, three-dimensional network of bundles with characteristic mesh size of the order of the persistence length of F-actin.
The cell cytoskeleton comprises three negative charged filamentous proteins, which include, filamentous-actin with a 8.5 nm diameter, the intermediate filaments, 10 nm in diameter, and microtubules, 25 nm in diameter. The cytoskeleton is involved in a range of cell functions including mechanical stability, cell locomotion, intracellular trafficking and signal transduction. We will describe recent work on interactions and structures in supramolecular assemblies of the actin cytoskeleton under in vitro conditions.

We show in Fig. 1 (left) laser scanning confocal microscope images of the actin cytoskeleton in mouse fibroblast cells. (The image is showing only the section closest to the cover slip to which the cell is attached.) The actin cytoskeleton provides a structural framework for the mechanical stability of eukaryotic cells and is involved in functions including cell adhesion, motility, and division. Actin is found both in a monomeric globular (G) actin state and as polymerized filamentous (F) actin. Bundles, comprised of closely packed parallel arrangements of F-actin, and networks, containing F-actin crisscrossed at some large angle, form the most common known supramolecular structures in cells. Interactions between F-actin and distinct actin cross-linking proteins, may lead to two-dimensional (2D) networks and bundles of F-actin interacting with the plasma membrane to determine cell shape, or 3D networks of F-actin imparting gel-like properties to the cytosol.
In Figure (Fig. 1, left, broken arrow), actin bundles can be seen traversing the length of a cell near the center of the image. These bundles are components of stress fibers, which end in focal adhesion spots responsible for cell adhesion, for example, to the extracellular matrix in the space between cells in vivo. Radial bundles of lamellipodium in membrane-protrusions (Fig. 1, left, solid arrow) are also associated with adhesion complexes.
When filamentous actin is allowed to mix in-vitro with a-actinin, an actin cross-linking protein purified from cells, a remarkable new type of biologically inspired polymer-network is spontaneously formed with fundamentally new properties. Three-dimensional laser scanning confocal microscopy carried out in our laboratory at UCSB has revealed a network of bundles on the micron scale (Fig. 1, right). This is in contrast to the commonly observed network of single filaments observed in cells. The branching (bifurcation) of the bundles on this meso-scale is evident and leads to a well-defined mesh size (1).
In order to understand the interaction between actin and a-actinin which leads to this previously overlooked self-assembling system, small angle x-ray diffraction (SAXRD) experiments were performed on beam lines 4-2 and 10-2 at SSRL to probe the local structural organization within the bundles (1). Experiments were done for different a-actinin:G-actin molar ratios (g) at 100mM KCl concentration and an actin concentration of 2.5mg/ml. At g = 1:90 the x-ray diffraction signature is barely different from background, while at g = 1:50 a sharp increase in small angle x-ray scattering (SAXS) indicates the formation of macromolecular structures. The enhanced SAXS at g = 50 and g = 25 is indicative of the formation of a loose network of F-actin filaments (Fig. 2, left), but where the F-actin - F-actin correlations (observed for g ≤ 10) have not yet set in due to the low a-actinin concentration.
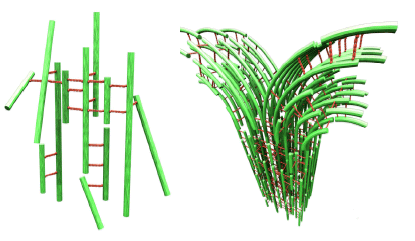
At g = 1:10 and above, broad diffraction peaks indicative of the onset of F-actin correlations are visible in the SAXRD. This data however reveals for the first time that the actin filaments are organized into a distorted (quasi) square lattice within the bundles with an approximately 30-nanometer unit cell dimension (Fig. 2, right). The peak widths obtained from SAXRD are consistent with a model of inherent disorder within the bundles where the width broadening scales as q2 consistent with short-range positional ordering. Figure 2 (right) demonstrates the ladder-like structure within the bundles at a branching site. It is proposed that the bundling process proceeds in a zipper-like fashion between adjacent fibers. At a bifurcation point an F-actin fiber will bend away from the main bundle where new fibers zip on creating a branch. The mechanical properties of this network of bundles should reveal length-scale dependent behavior. On the network mesoscale, elastomer-like elasticity of the network should be observed. On the small scales within the bundle 250 nm we expect a non-zero shear and compression moduli resulting from the finite-sized quasi-2D lattice within the bundles.
A separate motivation for the work described here was provided by our NIH funded research project within the group focused on developing optimal non-viral cationic lipid (CL) carriers of genes for gene delivery. Gene carriers based on lipids — rather than on engineered viruses — are among the most promising technologies for delivering genes into cells for gene therapy and therapeutics (2,5). Indeed, nearly one quarter of ongoing gene therapy clinical trials worldwide are conducted with non-viral methods including lipids, polymers, and naked DNA (http://www.wiley.co.uk/genetherapy/clinical/). Synchrotron x-ray diffraction work at SSRL by our group has solved the structures formed by CL-DNA complexes (6,7).
From a fundamental perspective, a full understanding of gene transfer technology by cationic lipids will emerge once the interactions between exogenous lipid and the biomolecules of the cell are understood. Work by our group at SSRL, which studied the interactions between CLs and F-actin in-vitro, showed that CL-actin complexes may spontaneously assemble into multilamellar tubules (8). The study reported here constitutes an important step at elucidating the structure of F-actin/a-actinin complexes, which will be followed by future experiments probing the interactions between F-actin/a-actinin and CL-DNA complexes mimicking gene delivery conditions. Thus, one may expect that actin filaments may dissociate from bundles in the cell cytoplasm and interact with and remove cationic lipids from CL-DNA complexes, thus, facilitating DNA release from complexes in cells for expression of genes.
Acknowledgements The work described in this report was supported by the National Institutes of Health grant GM-59288, the National Science Foundation, grants DMR-0203755 and CTS- 0103516, and the Department of Energy's Office of Basic Energy Sciences under contract number W-7405-ENG-36 with the University of California. X-ray diffraction was conducted at The Stanford Synchrotron Radiation Laboratory supported by the US Department of Energy.
- "Structure of Actin Cross-Linked with a-Actinin: A Network of Bundles", O. Pelletier, E. Pokidysheva, L. S. Hirst, N. Bouxsein, Y. Li, C. R. Safinya Physical Review Letters 91 (14), 148102-1-4 (2003). (Cover Image for October 3 issue).
- "Pharmaceutical Perspectives of Nucleic Acid-Based Therapeutics", Eds. Ram I. Mahato and Sung Wan Kim (Taylor & Francis, London & New York 2002).
- "Structures of Lipid-DNA Complexes: Supramolecular Assembly and Gene Delivery", C. R. Safinya, Current Opinion in Structural Biology 11 (4) 440-448 (2001).
- "Breaching the Membrane", in Science, News Focus: Drug Delivery: Breaching the Membrane,by J. Alper, 296, 838-839 (2002).
- "Gene Delivery -Without Viruses", in Chemical & Engineering News, Cover Story, by Celia M. Henry, pp. 35-41, November 26, 2001.
- "An Inverted Hexagonal Phase of DNA-Cationic Liposome Complexes Related to DNA Release and Delivery", I. Koltover, T. Salditt, J.O. Raedler, C. R. Safinya, Science 281, 78- 81 (1998).
- "Structure of DNA-Cationic Liposome Complexes: DNA Intercalation in Multi-Lamellar Membranes in Distinct Interhelical Packing Regimes", J. O. Raedler, I. Koltover, T. Salditt, C. R. Safinya Science 275, 810-813 (1997).
- "Hierarchical Self-Assembly of F-Actin and Cationic Lipid Complexes: Stacked 3-Layer Tubule Networks", G. C. L. Wong, J. X. Tang, A. Lin, Y. Li, P. A. Janmey, C. R. Safinya Science, 288 2035-2039 (2000).
O. Pelletier, E. Pokidysheva, L. S Hirst, N. Bouxsein, Y. Li and C. R Safinya, "Structure of Actin Cross-Linked with α-Actinin: A Network of Bundles", Phys. Rev. Lett. 91, 148102 (2003)




