Riboswitches are non-coding (nc)RNA elements typically found in the 5¢ leader sequences of bacterial messenger RNAs where they control the expression of a downstream gene in response to levels of a cognate cellular metabolite. Metabolite binding to riboswitch’s aptamer domain triggers a conformational change that alters the accessibility of a gene-regulatory sequence known as the expression platform1. Riboswitches are believed to participate in biochemical feedback loops, often controlling genes essential for fitness and survival. One of the best-studied riboswitches is the class I prequeuosine1 (preQ1-I) riboswitch, which is broadly distributed in the biosphere. PreQ1-I riboswitches are typically 30-35 nucleotides long2 and represent a powerful platform to study RNA structure3, 4, dynamics5, 6 and small molecule-RNA interactions7, 8. This riboswitch regulates the biosynthesis of queuosine (Fig. 1a), the loss of which has been linked to reduced bacterial virulence9, suggesting that these small riboswitches are viable antimicrobial targets.
Although over 55 riboswitch classes have been discovered, only a handful recognize more than one metabolite10. Recently, the Wedekind Lab at the University of Rochester School of Medicine and Dentistry structurally characterized a preQ1-I riboswitch that senses two preQ1 metabolites (Fig. 1b). Whereas all known multi-metabolite sensing riboswitches position their ligands in separate binding pockets, this small riboswitch recognizes its two ligands in a single binding pocket using novel interactions.
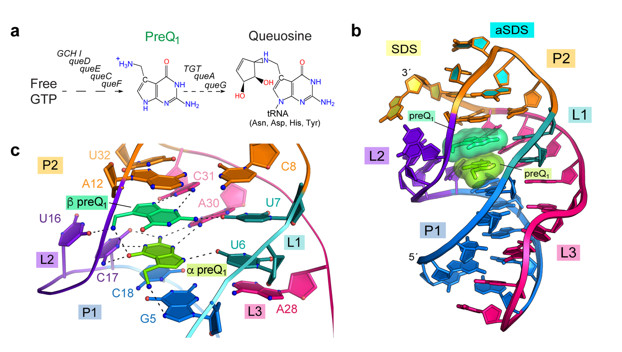
Wedekind and co-workers discovered the 34-nucleotide preQ1-I riboswitch in the genome of Carnobacterium antarcticus using an NCBI BLAST search. The riboswitch crystallized readily, and x-ray diffraction data were collected to 2.60 Å resolution on BL12-2, which revealed a pseudoknotted fold that engulfs two stacked metabolites. In addition to specific RNA interactions, the metabolites interact with each other via hydrogen bonding and aromatic stacking (Fig. 1c). The structure revealed key nucleotides that participate in preQ1 recognition, which were validated experimentally by isothermal titration calorimetry and bacterial reporter assays in live cells. This work revealed that the two metabolites bind with positive cooperativity and that both preQ1 molecules must bind for effective gene regulation.
The team also demonstrated that the mode of dual, stacked metabolite recognition is a hallmark of the most dominant preQ1-sensing riboswitch class in terms of its presence in known bacterial DNA sequences. Moreover, existing riboswitches can provide insight into the capabilities of catalytic RNAs (ribozymes) that might have evolved in a prebiotic RNA world to carry out metabolism. The high conservation of this riboswitch across multiple bacterial phyla suggests that it has ancient origins. The team hypothesizes that this small aptamer could have arisen from a now-extinct ribozyme that held two metabolites close together for covalent bond formation or to exchange a chemical group. This finding changes our view of how RNA can interact with ligands and suggests a sophistication akin to single-domain proteins that bind multiple substrates in a one pocket.
Looking toward the future, preQ1-I riboswitches are utilized by numerous bacteria to maintain homeostasis, including the antibiotic resistant pathogen Nesseria. gonorrheae11. The newly characterized, dual-metabolite binding pocket opens new opportunities to develop high-specificity small molecules that target this important gene-regulatory RNA, while averting cross-reactivity with the targets of natural metabolites.
R. R. Breaker, “Riboswitches and Translation Control”, Cold Spring Harb Perspect Biol 10:a032797.
A. Roth et al., “A Riboswitch Selective for the Queuosine Precursor PreQ1 Contains an Unusually Small Aptamer Domain”, Nat. Struct. Mol. Biol. 14, 308-317 (2007).
R. C. Spitale, A. T. Torelli, J. Krucinska, V. Bandarian and J. E. Wedekind, “The Structural Basis for Recognition of the PreQ0 Metabolite by an Unusually Small Riboswitch Aptamer Domain”, Journal Biol Chem 284, 11012-11016 (2009).
J. L. Jenkins, J. Krucinska, R. M. McCarty, V. Bandarian and J. E. Wedekind, “Comparison of a PreQ1 Riboswitch Aptamer in Metabolite-bound and Free States with Implications for Gene Regulation”, J. Biol. Chem. 286, 24626-24637 (2011).
A. J. Rinaldi, P. E. Lund, M. R. Blanco and N. G. Walter, “The Shine-Dalgarno Sequence of Riboswitch-regulated Single mRNAs Shows Ligand-dependent Accessibility Bursts”, Nat. Commun. 7, 8976 (2016).
K. C. Suddala et al., “Single Transcriptional and Translational PreQ1 Riboswitches Adopt Similar Pre-folded Ensembles that Follow Distinct Folding Pathways into the Same Ligand-bound Structure”, Nucleic Acids Res. 41, 10462-10475 (2013).
E. Neuner, M. Frener, A. Lusser and R. Micura, “Superior Cellular Activities of Azido- over Amino-functionalized Ligands for Engineered PreQ1 Riboswitches in E. coli”, RNA Biol. 15, 1376-1383 (2018).
C. M. Connelly et al., “Synthetic Ligands for PreQ1 Riboswitches Provide Structural and Mechanistic Insights into Targeting RNA Tertiary Structure”, Nat. Commun. 10, 1501 (2019).
J. K. Hurt, S. Olgen and G. A. Garcia, “Site-specific Modification of Shigella flexneri virF mRNA by tRNA-guanine Transglycosylase In vitro. Nucleic Acids Res. 35, 4905-4913 (2007).
P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, “Riboswitch Diversity and Distribution”, RNA (New York, NY) 23, 995-1011 (2017).
CDC. Biggest Threats and Data: 2019 Antibiotic Resistance Threats Report. Center for Disease Control and Prevention (2019).
G. M. Schroeder, C. E. Cavender, M. E. Blau, J. L. Jenkins, D. H. Mathews and J. E. Wedekind, “A Small RNA that Cooperatively Senses Two Stacked Metabolites in One Pocket for Gene Control”, Nat. Commun. 13, 199 (2022) doi: 10.1038/s41467-021-27790-8




