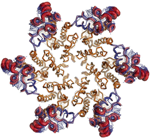
A Crystal Structure of HIV-1 Capsid Proteins Shows Flexibility Necessary for
Capsid Assembly
summary written by Raven Hanna
The genome of the human immunodeficiency virus (HIV-1) is bundled inside a capsid composed of about 1,500 copies of the viral Capsid Assembly (CA) protein. These proteins first assemble into substructures, each with six proteins, and these substructures come together to create the cone-shape casing of the virus. Disruption of capsid formation is a natural target for HIV therapies, and knowing the atomic structure of the CA proteins in the capsid would be useful for drug development. However, inherent flexibility in these molecules makes obtaining quality crystals difficult.
A team of researchers led by Mark Yeager from The Scripps Research Institute have solved the crystal structure of the hexameric CA protein substructure at 2.7 Å. To obtain their crystals, they used more stable subunits obtained by crosslinking the proteins at sites suggested by a 9 Å electron microscopy structure. They collected data using SSRL Beam Line 7-1 and solved the structure using molecular replacement. Their structure shows that the N-terminal domains are packed tightly in the center of the hexameric structure, but the C-terminal domains on the outside have more structural variability. This flexibility is probably necessary to allow the hexamers to assemble into the cone-like shape.
This research will aid discoveries of new capsid-targeted therapeutic agents
against HIV-1 using structure-based drug design. This work was published in the
June 11, 2009, issue of Cell.
To learn more about this research see the full Scientific Highlight
Pornillos, O., Ganser-Pornillos, B. K., Kelly, B. N., Hua, Y., Whitby, F. G., Stout, C. D., Sundquist, W. I., Hill, C. P., and M. Yeager, M. (2009). X-ray structures of the hexameric building block of the HIV capsid, Cell 137, 1.