

The ribonucleoprotein genomic complex of human immunodeficiency virus type 1
(HIV-1) is encased within the mature capsid, a predominantly cone-shaped shell
assembled from ~1,500 copies of the viral CA protein [1].
Packaging of the viral genome and its associated enzymes into the capsid is
required for their delivery into host cells. To form a closed shell the CA
protein assembles into ~250 hexamers. The CA hexamer consists of a ring of six
N-terminal domains around a central core, surrounded by an outer ring of six
more mobile C-terminal domains. Based on crystal structures of the individual
domains, and electron microscopy (EM) data of assembled hexamers, the
C-terminal domains are in contact with neighboring N-terminal domains by
rotation of 60° about the hexamer 6-fold axis. Flexibility between these
domains allows the CA hexamer to subtly adapt its shaped within the curved
array of the assembled capsid.
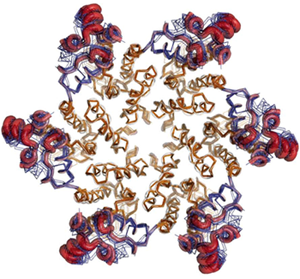
Figure 1. Superposition of multiple copies of the HIV-1 CA
hexamer, showing plasticity of the smaller C-terminal domain (blue lines and
red tubes) relative the N-terminal domain (brown). The C-terminal domain of
one CA subunit interacts with the N-terminal domain of an adjacent CA subunit
by clockwise rotation about the central 6-fold axis. Variability in the
conformation and position of the C-terminal domains allows the hexamer to
assembly into the curve, cone-shaped capsid of the virus.
Until now it has not been possible to crystallize the intact CA hexamer. By
modeling crystal structures of the individual CA domains into a 9 Å
resolution EM map of assembled hexamers [2], ten possible
disulfide crosslinks were designed to stabilize interactions between adjacent
N-terminal domains. Engineering of the double mutants and biochemical assay
identified one crosslinked hexamer as being uniquely stable and homogeneous.
This stabilized hexamer yielded large single, orthorhombic crystals diffracting
to 2.7 Å at SSRL beam line 7-1; the structure was solved by molecular
replacement with reference to the EM model. The crystals contained two copies
of CA hexamers per asymmetric unit, providing 12 independent views of
previously unknown details for the interface between N- and C-terminal domains
(PDB deposition 3H4E). Analysis of the structure revealed that networks of
water molecules mediate interactions between hydrogen bonding residues and
sites on helices of each domain, allowing the distinct variability required for
capsid formation. At the same time, comparison showed that the C-terminal
domain exhibits internal plasticity in the positioning of its helices
(Fig. 1).
The structure also afforded high resolution views of the packing of N-terminal
domains around the hexamer 6-fold axis, and of the 2-fold interaction between
C-terminals domain as they occur in the capsid.
The new crystal structure permits assembly of the intact HIV-1 capsid to be
visualized at atomic resolution. Because the capsid performs an essential role
early in the viral replication cycle, inhibition of capsid assembly by small
molecules is a new therapeutic strategy for the treatment of HIV/AIDS. To date
all FDA approved drugs for AIDS target the viral enzymes protease, reverse
transcriptase, and integrase. The N- and C-terminal domains are attractive
sites for inhibition because experimental inhibitors of HIV-1 capsid assembly
target them [3, 4]. New classes of drugs that target the interface of the N-
and C-terminal domains to inhibit capsid assembly, or alternatively, enhance
capsid stability and prevent uncoating, would provide novel means to intervene
in the virus replication cycle, and important tools to combat the evolution of
resistance in HIV. The mode of binding of such compounds can now be studied in
detail, and new binding sites can be identified, facilitating structure-based
drug design strategies.
This study was funded by NIH grants R01-GM066087, P50-GM082545, and the George
E. Hewitt Foundation for Medical Research.
Primary Citation
Pornillos, O., Ganser-Pornillos, B. K., Kelly, B. N., Hua, Y., Whitby, F. G.,
Stout, C. D., Sundquist, W. I., Hill, C. P., and M. Yeager, M. (2009). X-ray
structures of the hexameric building block of the HIV capsid, Cell 137, 1.
References
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.