Identifying Novel Targets that Extend the Effectiveness of HIV Protease
Inhibitors
summary written by Raven Hanna
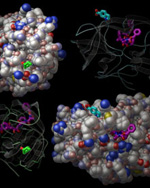
HIV protease is a common and critical drug target for combating HIV infection and AIDS. As HIV develops resistance to anti-viral drugs, new therapies are required. Since most of the virus's mutations that confer drug resistance cluster in the active site of the protease, scientists are interested in molecules that may bind other places on the enzyme. Computer simulations aid the design of drugs and fragments, which are smaller than typical drugs, to bind the enzyme's surface in a way that compliments the activity of traditional active-site binding drugs.
A team of scientists led by Prof. Dave Stout at The Scripps Research Institute has used SSRL beam lines to crystallographically screen fragment binding to HIV protease. They screened 400 fragments and evaluated 800 crystals using SSRL's high-throughput robotic sample automounter system. They found two novel surface-binding sites that induce conformational changes in the protease.
Their study shows that applying this high-throughput method can identify new
potential drugs and drug targets that work in combination with existing drug
therapies. This may allow current drugs to continue being effective, despite
viral mutations. This work was published in the March 2010 issue of
Chemical Biology and Drug Design.
To learn more about this research see the full Scientific Highlight
Perryman, A. L., Zhang, Q., Soutter, H. H., Rosenfeld, R., McRee, D., Olson, A. J., J. E. Elder, J. E. & Stout, C. D. (2010) Fragment-based Screen against HIV Protease. Chem. Biol. Drug Des. 75: 257-268.