
 The translation of the genetic code into proteins relies on accurate ligation
of amino acids to their matching tRNAs, by a group of enzymes known as
aminoacyl-tRNA synthetases. Mistranslation from confusing (by alanyl-tRNA
synthetase (AlaRS)) the amino acids serine or glycine for alanine has profound
pathological consequences, both in mammals1 and
bacteria2. To achieve accurate translation, AlaRSs
in all species have an aminoacylation site and an editing site that work
collaboratively to hydrolyze the misacylated Ser/Gly-tRNAAla. A
third domain, C-Ala domain, is universally tethered to the end of the editing
domain in AlaRSs throughout the 3 kingdoms of life, but heretofore had no known
function. In a work recently published in Science, a research team led by
Profs. Paul Schimmel and Xiang-Lei Yang at The Scripps Research Institute
determined the 3-D structure and the function of C-Ala and showed how these
results, in turn, shed light on the early evolution of the apparatus for
accurately translating the genetic code3.
The translation of the genetic code into proteins relies on accurate ligation
of amino acids to their matching tRNAs, by a group of enzymes known as
aminoacyl-tRNA synthetases. Mistranslation from confusing (by alanyl-tRNA
synthetase (AlaRS)) the amino acids serine or glycine for alanine has profound
pathological consequences, both in mammals1 and
bacteria2. To achieve accurate translation, AlaRSs
in all species have an aminoacylation site and an editing site that work
collaboratively to hydrolyze the misacylated Ser/Gly-tRNAAla. A
third domain, C-Ala domain, is universally tethered to the end of the editing
domain in AlaRSs throughout the 3 kingdoms of life, but heretofore had no known
function. In a work recently published in Science, a research team led by
Profs. Paul Schimmel and Xiang-Lei Yang at The Scripps Research Institute
determined the 3-D structure and the function of C-Ala and showed how these
results, in turn, shed light on the early evolution of the apparatus for
accurately translating the genetic code3.
 |
Figure 1. Structure of the C-Ala domain of Aquifex aeolicus AlaRS indicates it has a single-stranded nucleic acid binding fold with a tRNA binding surface located at one side. |
Distinct from all the other aminoacyl-tRNA synthetases, AlaRSs have a group of
free-standing partners-AlaXps-that are wide-spread in the three
kingdoms4.
AlaXps specifically hydrolyze the tRNAAla's that has been mischarged with
serine instead of alanine. This activity provides functional redundancy to the
editing activity that is imbedded in AlaRSs5,6,7. The AlaXps are homologous to
the editing domains of AlaRSs and some of them are also fused to C-Ala.
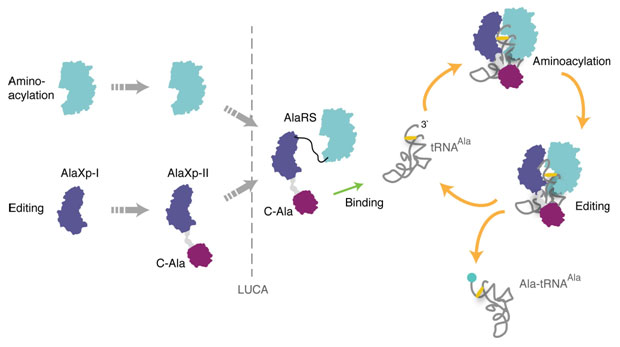
Figure 2. Proposed assembly of AlaRS in evolution. The shorter
free-standing editing protein (AlaXp-I) is proposed to have coexisted in trans
with the free-standing aminoacylation domain of AlaRS. The longer AlaXp-II
formed by combining AlaXp-I with the C-Ala domain, which was able to bring
together editing and aminoacylation centers to create the present-day AlaRS.
Using data collected at SSRL Beam line 11-1, the team quickly determined the
three dimensional structure of C-Ala by the method of single anomalous
diffraction. The automatic crystal-mounting at SSRL enabled the efficient
screening and data-collecting of the crystals, just weeks after the cloning of
this domain. The x-ray structure of C-Ala at 1.85 angstroms revealed a
single-stranded nucleic acid binding fold, which was seen in some DNA
exonucleases (such as RecJ) (Fig. 1). Site-specific footprinting showed that
C-Ala binds to the elbow region of tRNAAla. Other experimental data, including
binding and kinetic analysis, showed that C-Ala is a major tRNA-binding module
of AlaRS, and serves as a bridge to collaboratively join editing with
aminoacylation on one tRNA3. Separately, phylogenetic analysis showed that
AlaXp evolved in the ancient community and that the imbedded editing domain of
AlaRS originated from AlaXp (Fig. 2). Thus, C-Ala may have also had the
historical role of being the bridge that first joined editing with
aminoacylation.
Primary Citation
Min Guo, Yeeting E. Chong, Kirk Beebe, Ryan Shapiro, Xiang-Lei Yang and Paul
Schimmel "The C-Ala domain brings together editing and aminoacylation functions
on one tRNA". Science, 325, 744-747 (2009).
References
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.