
 Modular polyketide synthases (PKSs) such as the 6-deoxyerythronolide B synthase
(DEBS) are a large family of polyfunctional, multi-subunit enzymes that
catalyze the biosynthesis of structurally complex and medicinally important
natural products known as polyketides (1, 2).
Although polyketides are biosynthesized from simple acyl-CoA building blocks,
their structural complexity often precludes the development of practical
laboratory synthetic routes, leaving fermentation as the only viable source
for the commercial production of these pharmaceutically and agriculturally
useful agents. As the model system of modular PKSs, DEBS has been studied
genetically and biochemically over the past two decades.
Modular polyketide synthases (PKSs) such as the 6-deoxyerythronolide B synthase
(DEBS) are a large family of polyfunctional, multi-subunit enzymes that
catalyze the biosynthesis of structurally complex and medicinally important
natural products known as polyketides (1, 2).
Although polyketides are biosynthesized from simple acyl-CoA building blocks,
their structural complexity often precludes the development of practical
laboratory synthetic routes, leaving fermentation as the only viable source
for the commercial production of these pharmaceutically and agriculturally
useful agents. As the model system of modular PKSs, DEBS has been studied
genetically and biochemically over the past two decades.
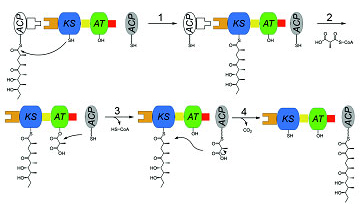 |
Figure 1. Chain elongation cycle catalyzed by a PKS module. |
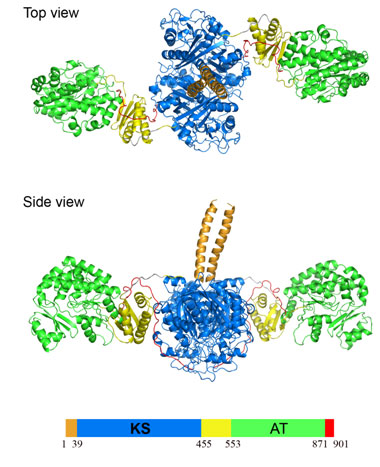
Using x-ray diffraction data collected at SSRL Beam Line 11-1, Chaitan Khosla's
group at Stanford University has determined the crystal structure of a 194 kDa
homodimeric fragment of the KS-AT didomain of DEBS module 5 (Figure 2). This
high resolution structure of the first didomain structure of a PKS module
provides insights into the complex structural organization of the modular
polyketide synthases machinery. The didomain structure was solved by
multiwavelength anomalous dispersion techniques and has 40908 atoms (582 kDa)
per asymmetric unit. To our knowledge this is the largest x-ray crystal
structure that has been solved to date using the MAD technique. This project
required the screening of around 600 crystals using SSRL's SAM robot.
Figure 2.
Structure of the KS-AT didomain from DEBS module 5. The KS-AT protein forms a
homodimer.
The 194 kDa homodimeric fragment contains full-length KS and AT domains as well
as three flanking linkers: the N-terminal linker, an intervening KS-to-AT
linker, and post-AT linker. The structure reveals that the KS domain adopts an
abab fold, the AT domain contains an
a, b-hydrolase-like core
domain and an appended smaller ferredoxin-like subdomain. The three linkers are
also structurally well defined. The N-terminal linker forms a coiled-coil
structure, the KS-to-AT linker is formed by a three stranded b-sheet packed against two a-helices on one side, representing a protein fold not
previously reported in the Protein Data Bank, the post-AT linker wraps back
over both the AT domain and the KS-to-AT linker so as to interact specifically
with the KS domain. Both the KS-to-AT linker and the post-AT linker play
important structural roles in fixing the relative positions of the KS and AT
didomain. The crystal structure also reveals that the active site Cys199
residue of the KS domain is more than 80 Å away from the active site Ser642
residue of the AT domain. This distance is too large to be covered simply by
alternative positioning of statically anchored, fully extended
phosphopantetheine arm of the ACP domain. Thus, substantial domain
reorganization may be necessary for the ACP to interact successively with both
the AT and the KS domains of this prototypical polyketide synthase module.
These findings emphasize the critical role of unconserved but structurally well
defined linkers as well as large interdomain movements in the structure and
function of these remarkable modular megasynthases. The 2.7-Å KS-AT structure
is fully consistent with a recently reported lower resolution, 4.5-Å model of
fatty acid synthase structure (4), and emphasizes the close biochemical and
structural similarity between polyketide synthase and fatty acid synthase
enzymology.
In addition to providing the first atomic-level glimpses into the core
catalytic domains of multi-modular PKSs, this prototypical structure also
presents a fundamentally new perspective for future biochemical and engineering
investigations into this remarkable family of modular megasynthases.
This work was supported by NIH grants CA 66736 and GM 22172
Primary Citation
References
Tang, Y., Kim, C.-Y., Mathews, I. I., Cane, D. E., Khosla, C. (2006) The 2.7Å
Structure of a 194-kDa Homodimeric Fragment of the 6-Deoxyerythronolide B
Synthase. PNAS, 103, 11124-11129.
| SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. |