
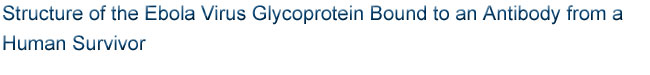
Ebolavirus: The ebolavirus causes a severe hemorrhagic fever with 50-90%
lethality for which no vaccines or treatments are yet available. The more
frequent re-emergence of the virus, its high prevalence among wildlife, and
ease of importation of the virus make it a significant public health concern. A
team of researchers have recently determined the crystal structure of the oligomeric, viral
surface glycoprotein in complex with a rare antibody derived from a human
survivor. This work explains how the glycoprotein, termed GP, mediates host
recognition, drives fusion of the viral and host membranes and masks itself
from immune surveillance. The structure also explains why antibodies that
neutralize the virus are so rare, identifies the very few sites to which a
neutralizing antibody might bind, and thus, provides templates for vaccines and
antibodies against the virus.
The glycoprotein GP is the sole resident of the ebolavirus surface and is
responsible for attaching to and entering new host cells, shielding of the
viral surface from immune surveillance, and maintenance of viral stability
between hosts (often in caves for long periods of time). Determination of the
crystal structure of GP was critical for understanding these processes and in
design and improvements of vaccines and therapeutics. However, structures of
Structural arrangement and rearrangement: Ebolavirus GP is cleaved by furin to
yield two subunits termed GP1 and GP2 with separate structural and functional
roles. Of these, GP1 is responsible for receptor engagement while GP2 mediates
fusion of viral and host membranes. The crystal structure illustrates that the
450 kDa GP forms a three-lobed chalice shape with the bowl of the chalice
assembled by the three GP1 subunits (Figure 1). The stem of the chalice is
formed by three GP2 subunits that cradle and encircle the GP1 trimer. Here, the
internal fusion loop and heptad repeat region of GP2 together wrap around GP1,
and in turn, hydrophobic residues of GP1 clamp the heptad repeat of GP2 into
its metastable, prefusion conformation (Figure 2). This clamp is released in
entry through an as yet unidentified process, allowing GP2 to spring into its
more stable, six-helix bundle conformation and trigger fusion of virus and host
membranes.
Insight into receptor binding and entry: This structure, the first
near-complete structure of any filovirus glycoprotein, allowed identification
of a putative receptor-binding site on GP. This site is sequestered in the bowl
of the GP trimer, further masked by a novel glycan cap domain and a heavily
glycosylated, unstructured mucin-like domain. GP was known to be cleaved by
cathepsin proteases as an essential step in entry, but the precise site or role
of cleavage was unknown. Importantly, the crystal structure identifies the
probable cleavage site of GP and illustrates how cleavage at this site uncaps
the receptor binding regions freeing them for interaction with host cell
receptor(s). Thus, the crystal structure of GP suggests that initial cellular
attachment occurs via interactions of cell surface lectins with the mucin-like
domain or other glycosylated regions on GP, and that the receptor-binding site
is revealed later in the endosome upon proteolytic processing.
Templates for vaccines and immunotherapeutics: The crystal structure also
reveals that most of GP is shielded by a thick cloak of carbohydrate and
identifies the very few sites left exposed and available for antibody binding.
Hence, this structure is now serving as a template for vaccines and antibodies
to target these newly revealed slits in ebolavirus's cloak.
Primary Citation:
References:
viral glycoproteins
in their native, viral surface forms can be difficult to
achieve as they are oligomeric, metastable, and heavily glycosylated. Indeed,
half or more of the molecular weight of ebolavirus GP is comprised of
heterogeneous carbohydrate and unstructured polypeptide. Through production of
some 140 versions of the viral glycoprotein, alone or in complex with one of
seven different antibodies, Jeffrey Lee, Marnie Fusco and Erica Ollmann Saphire of The
Scripps Research Institute were able to crystallize the trimeric, prefusion
form of GP, in complex with a neutralizing antibody derived from a human
survivor of the 1995 Kikwit, Zaire outbreak. The researchers had to grow
~50,000 crystals for this project, and screen the 800 largest crystals over ~30
trips to ALS and SSRL in order to find one crystal that would diffract to
3.4 Å (collected on ALS BL5.02) and permit structure determination. Importantly, the GP crystallized retains
all regions required for attachment, fusion and entry. Viruses pseudotyped with
the crystallization construct plus the transmembrane domain are functional in
infectivity assays and exhibit antibody neutralization profiles identical to
wild-type GP.
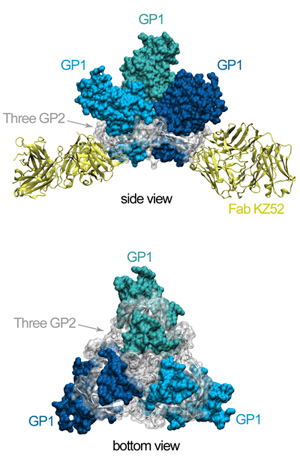
Figure 1. The crystal structure of ebolavirus GP reveals a three-lobed
chalice-like structure. The three GP1 subunits (colored blue and green),
mediate attachment to new host cells, and are tethered together by the three
GP2 subunits (white). GP2 forms the protein machinery which drives fusion of
the viral membrane with the host cell. The human antibody KZ52 (yellow) binds
an epitope at the base of the GP chalice where it bridges GP1 to GP2.
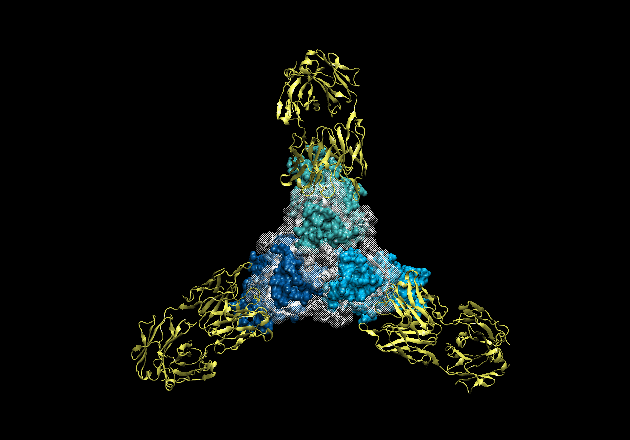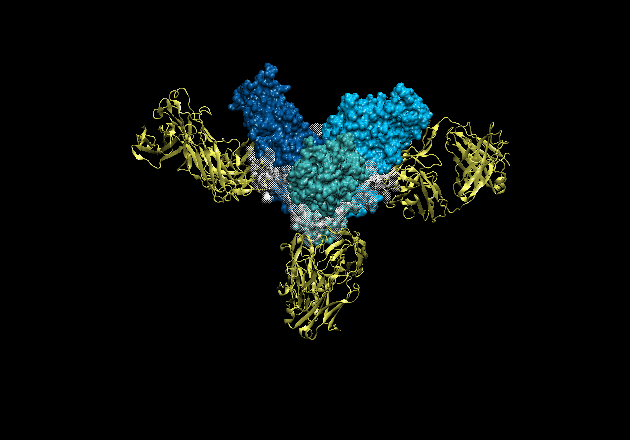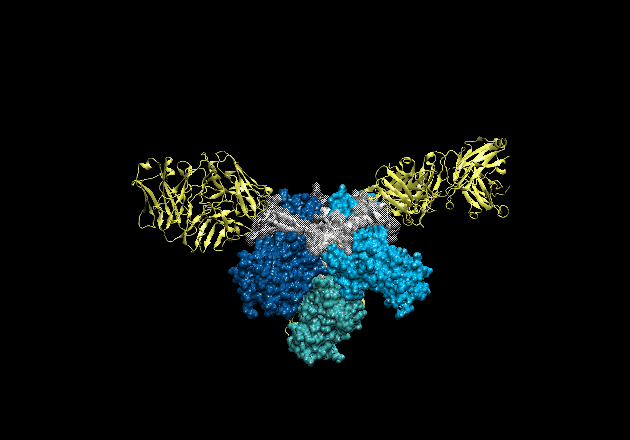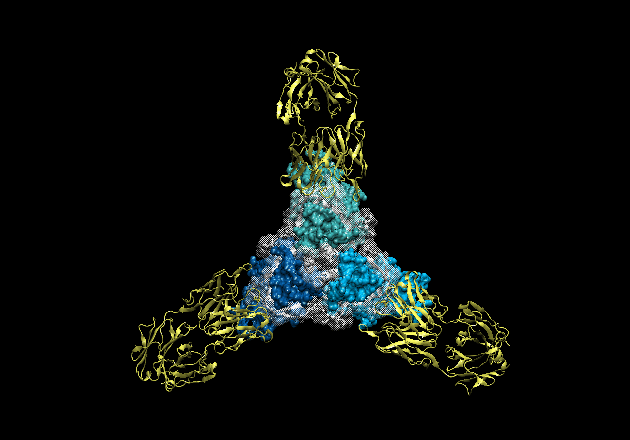
Figure 2. Movie of the rotating GP-KZ52 complex. Here, one can see how the
GP2 subunits (white) are wound around GP1 (blues) like thread around a spool.
GP1 forms a hydrophobic clamp on GP2, holding it in this metastable, prefusion
conformation on the viral surface.
Lee, J.E., Fusco, M.L., Oswald, W.B., Hessell, A.J., Burton, D.R., and Saphire,
E.O. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from
a human survivor. Nature, 454:177-182.
Lee, J.E.; Fusco, M.L.; Hessell, A.J.; Oswald, W.B.; Burton, D.R. and Saphire,
E.O. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from
a human survivor. Nature. 454:177-182.
Funding:
The authors would like to thank the NIH (EOS), the Burroughs Wellcome Fund
(EOS), and the Canadian Institute for Health Research (JEL) for funding.
For SSRL:
We greatly appreciated the ready and repeat access to SSRL beamlines to
screen the hundreds of crystals required to find one that would diffract
well.
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.