Revealing a Structural Weakness of the Deadly
Ebolavirus
summary written by Brad Plummer, SLAC
Communication Office
Scientists are one step closer to conquering the deadly Ebolavirus, thanks to research conducted at SSRL structural biology Beam Lines 9-2 and 11-1 and ALS Beam Line 5.02 by a team of researchers led by Erica Ollmann Saphire from The Scripps Research Institute. The results were published in the July 10 edition of the journal Nature.
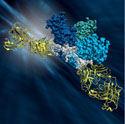
Using macromolecular crystallography techniques, the team solved the structure of a protein on the Ebolavirus's surface, called glycoprotein GP, in complex with a rare antibody identified in a human survivor. The glycoprotein-antibody complex proved especially challenging to crystallize and subsequently, to yield well-diffracting crystals. The team grew ~50,000 crystal samples and screened 800 of the largest, using in part the highly-automated robotics hardware and software at the SSRL beam lines, before finding a sample that would diffract to 3.4 Angstroms.
The Ebolavirus causes a severe hemorrhagic fever with 50-90% lethality for which no vaccines or treatments are yet available. In order for Ebolavirus to infect a host, it must first recruit one of the host's own enzymes to uncover a portion of the glycoprotein required for infection. This work explains how GP reveals its host binding sites, drives fusion of the viral and host membranes, and disguises itself from surveillance of the immune system. The structure also explains why antibodies that neutralize the virus are so rare, identifies the very few locations on the Ebolavirus where an antibody might attach itself, and thus provides structural templates that can be used for developing vaccines and antibodies against the virus.
To learn more about this research see the full Scientific Highlight
Lee, J.E., Fusco, M.L., Oswald, W.B., Hessell, A.J., Burton, D.R., and Saphire, E.O. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature, 454:177-182.