

Targeting of newly synthesized membrane proteins to the endoplasmic reticulum
(ER) is an important cellular process. Most membrane proteins are recognized
and targeted co-translationally by the signal recognition particle (SRP). A
number of membrane proteins (eg. SNAREs, apoptosis factors, and protein
translocation components) are 'tail-anchored' by a single carboxy-terminal
transmembrane domain. Due to this topological constraint, these proteins are
not able to follow the SRP-dependent co-translational pathway that typifies
most integral membrane proteins. Instead, these proteins must find their
correct membrane for insertion post-translationally (1). The ATPase Get3 was
the first protein identified directly involved in TA targeting and is part of
the Get pathway (Guided Entry of Tail-anchored proteins) that also contains the
ER membrane proteins Get1/2 and the putative ribosome receptor proteins Get4/5
(2). Multiple studies have shown that Get3 binds directly to the hydrophobic
tail-anchors and, in conjunction with ribosome and ER factors, utilizes an ATP
cycle to bind and then release TA proteins at the ER membrane.
To get a detailed mechanistic view of how Get3 performs its important targeting
function a team at Caltech led by Bil Clemons used data collected at SSRL Beam
Line 12-2 to determine the three-dimensional structure of Get3/TRC40 (Fig. 1).
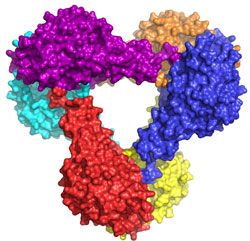
The structure of Get3 in the ADP form was a hexamer (Fig. 1a). It is expected
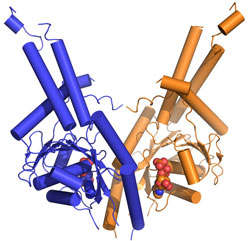
that Get3 functions as a dimer (Fig. 1b). Based on homology to other proteins,
the structure allowed for the prediction that binding of ATP would lead to a
dramatic conformational change bringing two hydrophobic grooves together to
form a TA-binding pocket. The team probed functional interfaces and essential
residues using phenotypic rescue allowing them to define a model of how Get3
couples ATP hydrolysis to the binding and release of TA-proteins.
This work represents the first structural study in this novel pathway. Future
work will allow for a refining of this model including the details of the
binding of substrate and activation for ATP hydrolysis.
Primary Citation
Suloway, C. J. M., Charton, J. W., Zaslaver, M., and Clemons, Jr. W. M. (2009)
Model for eukaryotic tail-anchored protein binding based on the structure of
Get3. PNAS, 106: 14849-14854.
References
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.