New Data from Beam Line 12-2 Reveals how Macromolecular Structural Distortions
Impact Function
summary written by Raven
Hanna
Biological macromolecules, like proteins and nucleic acids, are good examples of the form follows function paradigm; and, in the case of these molecules, deformation follows function as well. Flexibility in proteins and nucleic acids allows for the recognition of targets, the binding of complexes, and the adoption of functional configurations. Recent research at SSRL Beam Line 12-2 has revealed how distortion in macromolecular structure is linked to function. BL12-2 is the high-intensity, state-of-the-art undulator beam line for advanced macromolecular crystallographic studies funded by The Gordon and Betty Moore Foundation in cooperation with the California Institute of Technology.
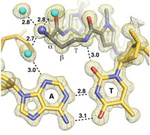
Research by a group led by Prof. Peter Dervan from California Institute of Technology shows how a small molecule compresses DNA's major groove upon binding to its minor groove. This structural distortion interrupts the recognition and binding of transcription factors, affecting gene expression. This work was published in the August 11, 2009, issue of the Proceedings of the National Academy of Sciences.
Chenoweth, D. M. and Dervan. P. B. (2009) Allosteric modulation of DNA by small
molecules PNAS,
106:13175-13179.
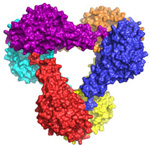
Another team, led by Bil Clemons from Caltech, solved several crystal structures of complexes of a transport protein (Get3). Get3 is responsible for delivering tail-anchored (TA) proteins to their proper locations after translation by the ribosome. Their data suggest a model in which both the binding of ATP and ATP hydrolysis cause dramatic conformational changes in Get3 that allow the binding and release of the TA-proteins. This work was published in the September 1, 2009, issue of the Proceedings of the National Academy of Sciences.
Suloway, C. J. M., Charton, J. W., Zaslaver, M., and Clemons, Jr. W. M. (2009)
Model for eukaryotic tail-anchored
protein binding based on the structure of
Get3. PNAS, 106:14849-14854.
To learn more about this research see the full Dervan and Clemons Scientific Highlights