
 Signals originating at the cell surface are conveyed by a complex system of
interconnected signaling pathways to the nucleus. They converge at
transcription factors, which in turn regulate the transcription of sets of
genes that result in the gene expression. Many human diseases are caused by
dysregulated gene expression and the oversupply of transcription factors may be
required for the growth and metastatic behavior of human cancers. Cell
permeable small molecules that can be programmed to disrupt transcription
factor-DNA interfaces could silence aberrant gene expression pathways.
Pyrrole-imidazole polyamides are DNA minor groove binding small molecules that
are programmable for a large repertoire of DNA motifs.
Signals originating at the cell surface are conveyed by a complex system of
interconnected signaling pathways to the nucleus. They converge at
transcription factors, which in turn regulate the transcription of sets of
genes that result in the gene expression. Many human diseases are caused by
dysregulated gene expression and the oversupply of transcription factors may be
required for the growth and metastatic behavior of human cancers. Cell
permeable small molecules that can be programmed to disrupt transcription
factor-DNA interfaces could silence aberrant gene expression pathways.
Pyrrole-imidazole polyamides are DNA minor groove binding small molecules that
are programmable for a large repertoire of DNA motifs.
Py/Im polyamides bind the minor groove of DNA sequence specifically
(1),
encoded by side-by-side arrangements of N-methylpyrrole (Py) and
N-methylimidazole (Im) carboxamide monomers. They have been shown to
permeate
cell membranes (2), access chromatin, and disrupt protein-DNA interactions.
X-ray crystallography of antiparallel 2:1 binding polyamides in complex with
DNA reveal a 1-2 Å widening of the minor groove (3). This modest
structural perturbation to the DNA helix by the side-by-side stacked
arrangement of aromatic rings does not explain the large number of
transcription factor-DNA interfaces disrupted by minor groove binding hairpin
Py/Im polyamides. It must be that the turn unit in the hairpin oligomer
connecting the 2 antiparallel strands plays a structural role.
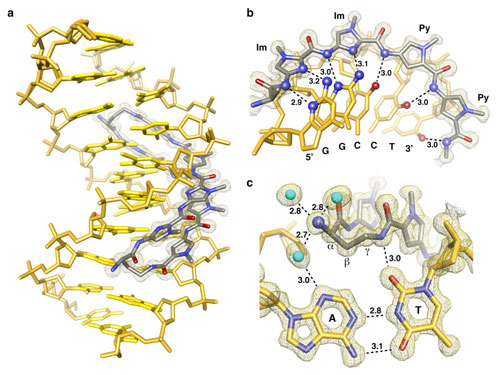
Figure 1
Using data collected at SSRL Beam Line 12-2, a research team led by Peter
Dervan from California Institute of Technology determined the three dimensional
high resolution an 8-ring cyclic Py/Im polyamide bound to the central 6 bp of
the sequence d(5'-CCAGGCCTGG-3')2 (Fig. 1). The structure reveals a 4 Å
widening of the minor groove and compression of the major groove along with a
>18° bend in the helix axis toward the major groove. This allosteric
perturbation of the DNA helix provides a molecular basis for disruption of
transcription factor-DNA interfaces by small molecules, a minimum step in
chemical control of gene networks. Allosteric control over transcription factor
regulatory networks by small molecules that bind distinct locations on promoter
DNA provides a mechanism for inhibiting excess transcription factor activity.
Primary Citation
Chenoweth, D. M. and Dervan. P. B. (2009) Allosteric modulation of DNA by small
molecules PNAS, 106:13175-13179.
References
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.