
Classical thymidylate synthases, encoded by the thyA and TYMS
genes, are present in most eukaryotes, including humans, and are frequently
targeted by chemotherapeutic and antibiotic drugs. A recently discovered class
of thymidylate synthases, the FDTSs encoded by the thyX gene has been
found primarily in prokaryotes and viruses including several pathogens and
biological warfare agents (see http://www.cdc.gov). Several organisms, including
human pathogens, rely solely on thyX for thymidylate synthesis. FDTSs share no
structure or sequence homology with classical thymidylate synthases (Fig 1),
and thus present a promising new frontier for antibacterial/antiviral drug
development.
|
 |
|
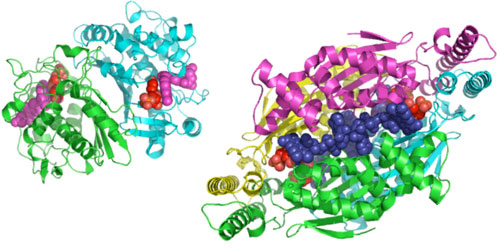
Figure 1. Structural comparison of classical TS and FDTS: Left: Ribbon
diagram of an E. coli TS (ecTS) dimer (PDB entry 2KCE). Both ligands are
highlighted as space filling shapes. The substrate dUMP is red and the cofactor
analogue (Zd1694, Ralitrexed), is magenta. Right: Ribbon diagram of a
tmFDTS tetramer (PDB entry 1O26). FAD (blue) and dUMP (red) are highlighted as
space filling shapes. It is clear that the active site is exposed to solvent
and that all four adenine rings interact at the center of the complex.
|
|
In an article published in Nature, research team lead by Prof. Kohen has
unraveled an unusual mechanism for the FTDS catalysis. A significant component
of the study involved structural data collected at SSRL Beam Line 9-2 by Dr.
Mathews following his successful crystallization of the wild type and two
critical mutants of FDTS. Other experimental components include kinetic and
isotopic analysis of the enzyme and mutants. The novel mechanism is an example
of thymidylate biosynthesis that occurs without an enzymatic nucleophile
(Scheme 1). The findings indicate that the putative active site nucleophile is
not required for FDTS catalysis, and no alternative nucleophilic residues
capable of serving this function can be identified. This study suggests that a
hydride is transferred from the reduced flavin cofactor directly to the uracil
ring, followed by an isomerization of the intermediate to form the product
thymidylate as illustrated in Scheme 1). The observations indicate a very
different chemical cascade than that of classical thymidylate synthases or any
other known biological methylation. The findings and chemical mechanism
proposed here, together with available structural data, suggest that selective
inhibition of FDTSs, with little effect on human thymine biosynthesis, should
be feasible. Because several human pathogens depend on FDTS for DNA
biosynthesis, its unique mechanism makes it an attractive target for antibiotic
drugs.
 |
|
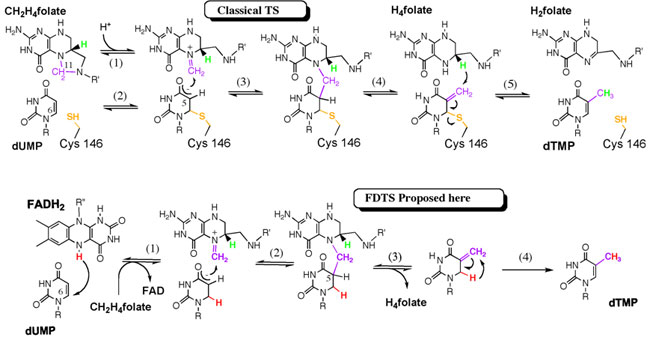
Scheme 1: Top, the chemical mechanism of classical TS. Bottom,
the newly proposed mechanism for FDTS. |
|
Primary Citation
Eric M. Koehn, Todd Fleischmann, John A. Conrad, Bruce A. Palfey, Scott A.
Lesley, Irimpan I. Mathews, and Amnon Kohen, "A Novel Chemical Mechanism
of Thymidylate Biosynthesis in Human Pathogens Containing the thyX Gene"
Nature 458, 919 (2009).
Related Press Release
UI Chemists' DNA Biosynthesis Discovery could Lead to Better Antibiotics,
University of Iowa News Release, April 15, 2009
http://news-releases.uiowa.edu/2009/april/041509biosynthesis_discovery.html
Biochemistry: Anchors Away, News and Views, Nature 458, 840-841 (16 April 2009)
doi:10.1038/458840a
References
|
Myllykallio, H. et al. An alternative flavin-dependent mechanism of thymidylate
synthesis. Science 297, 105-107 (2002).
Agrawal, N., Lesley, S. A., Kuhn, P. & Kohen, A. Mechanistic studies of a
flavin-dependent thymidylate synthase. Biochemistry 43, 10295-10301 (2004).
Mathews, I. I. et al. Functional analysis of substrate and cofactor complex
structures of a thymidylate synthase-complementing protein. Structure 11,
677-690 (2003).
|
|
| PDF version | | Lay
Summary | |
Highlights Archive
|
SSRL is supported
by the Department of Energy, Office of Basic Energy Sciences. The SSRL
Structural Molecular Biology Program is supported by the Department of Energy,
Office of Biological and Environmental Research, and by the National Institutes
of Health, National Center for Research Resources, Biomedical Technology
Program, and the National Institute of General Medical Sciences.
|
|



