

DNA is subject to multiple forms of damage that can occur either spontaneously,
through sources such as reactive oxygen species produced by cellular
metabolism, or through exogenous sources such as UV light and X-rays. Different
cellular pathways exist to detect and repair the distinct forms of damage, and
thus maintain the stability of the genome. The importance of the DNA repair
pathways is highlighted by inherited diseases that contain defects in pathway
components, resulting in marked increase in cancer incidence, rapid-aging
and/or neurological pathologies. One important family functioning in DNA repair
is the Rad60 family of proteins, with Rad60 being conserved from yeast to
humans. Sequence analysis of this family has revealed that members contain two
regions of similarity to SUMO, which have been termed SUMO-like domains (SLDs,
SLDs1 & 2). This inclusion of SLDs into a polypeptide chain is unusual because
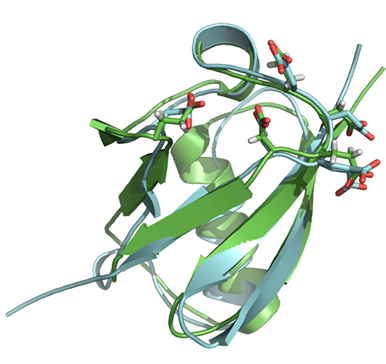
Figure 1. The 0.97 Å crystal structure of Rad60 SLD2.
The Rad60 SLD crystal structure depicted as cartoon, in green, structurally
superimposed onto human SUMO-1, in blue. The two structures share a common
fold, though many interaction motifs differ, except the E2 non-covalent
interface residues that are highlighted as sticks. Note that the ultra-high
resolution allowed for the refinement with hydrogen atoms, as indicated on
Rad60 SLD2 stick side chains.
Studies were conducted by a team of Structural Biologists, including Jeff Perry
and Andy Arvai, in the laboratory of John Tainer at The Scripps Research
Institute to define the roles of Rad60 proteins and their SLDs. Data was
collected on the C-terminal SLD2 of fission yeast (S. pombe) Rad60 at
SSRL beamline 11-1, which enabled structural determination to an ultra-high
0.97 Å resolution. This exceptional resolution allowed for a detailed
structural model that included hydrogen atoms in its final refinement (Fig. 1).
Interestingly, the Rad60 SLD2 backbone structure is well conserved with SUMO
(Fig. 1). Yet, the surface features differ between these proteins, suggesting
that most of the SUMO interactions with SUMO pathway components are not
conserved in Rad60. However, one exception is the conserved interface on SUMO
and on Rad60 SLD2 for a non-catalytic binding site on the SUMO E2 (Fig. 1);
this non-catalytic site is specifically used by the E2 to promote the formation
of SUMO chains (3,4,5). In
collaboration with Nick Boddy's yeast genetics laboratory, also at Scripps, a
structure-based mutation in Rad60 SLD2 was created that uncoupled its
interaction with the E2 partner on its chain-forming interface. Notably,
breaking this Rad60:E2 interaction resulted in a cellular hypersensitivity to
genotoxic stress, and also an increase in spontaneous recombination associated
with aberrant replication forks.
Overall, these results have provided the first detailed structural analysis of
SUMO-like domains, their interaction interfaces, and a mechanistic basis for
Rad60 functions in DNA-damage-responses, via interaction with the SUMO E2.
Understanding SUMO pathway interactions is of great significance as this
pathway is implicated in much human pathology, including cancer and
neurodegenerative disorders such as Alzheimer's, Parkinson's and Huntington's
diseases, and in viral infections.
Nick Boddy is a Scholar of the Leukemia and Lymphoma Society
(http://www.leukemia-lymphoma.org/hm_lls). This research was funded by US
National Institutes of Health grants GM068608 to Nick Boddy and GM081840
awarded to Nick Boddy and John Tainer.
Primary Citation
Prudden, J., Perry, J.J.P., Arvai, A.S., Tainer J.A., Boddy M.N. (2009).
Molecular mimicry of SUMO promotes DNA repair. Nature Struct. Mol. Biol.
16(5):509-16.
References
SUMO (Small Ubiquitin-like Modifier) is a small protein normally covalently
attached to target proteins, thereby altering target protein function and/or
cellular localization. The attachment of SUMO occurs via an enzymatic cascade
that uses three steps for ligation (1). The first step uses an
E1 enzyme that activates SUMO, the second step uses an E2 enzyme for
conjugation, and the third use an E3 protein for substrate recognition.
Multiple rounds of this SUMOylation cascade can also arise and this results in
the covalent attachment of SUMO chains, which appear to have roles in signaling
and in promoting the assembly of larger protein complexes (2).
Interestingly however, Rad60 SLDs lack a Gly-Gly motif that is present in SUMO
and is required for the activation and conjugation of SUMO to target proteins.
This suggests that Rad60 has instead novel interactions with SUMO pathway
components, potentially modulating the function of the SUMO pathway in DNA
repair and genome stability.
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.