
 |
Figure 1. Single crystal experimental setup on BL 9-3 at SSRL. Polarized
XAS of PS II single crystals were collected at 10 K using either a liquid He
cryostat or a liquid He cryostream (shown above). X-ray diffraction pattern was
collected using the imaging plate placed down stream of the crystal and was
used for orienting each crystal. |
Oxygen, that supports all aerobic life, is abundant in the atmosphere because
of its constant regeneration by photosynthetic water oxidation by green plants
and cyanobacteria. The metal catalyst responsible for this reaction resides in
a large protein complex, called photosystem II (PS II). The structure of the
catalytic Mn4Ca complex has been the subject of intense study ever
since Mn was identified as an essential element, using X-ray absorption, EPR,
and FTIR spectroscopies.1,2 In
addition, the four recent X-ray crystallography studies of PSII promise to add
valuable information to our knowledge about the structure of PS II and the
Mn4Ca complex.3-6 But
until now, the precise structure of the catalyst has eluded all attempts of
determination by these techniques, partly because of the susceptibility of the
Mn4Ca complex to X-ray radiation damage.7
Polarized extended X-ray absorption fine structure (EXAFS) measurements of
oriented PS II single crystals were collected by aligning the crystals
in situ
using the X-ray diffraction (XRD) pattern. The set up used for this study is
shown in Figure 1. EXAFS has the advantage of obtaining Mn-neighboring
atom distances with high accuracy and at a low X-ray dose. Moreover, the
polarized EXAFS spectra, collected using the pre-oriented single crystals using
XRD, provide a powerful filter for choosing among many proposed structural
models on the basis of the dichroism of the spectra from the single crystals.
These studies resulted in deriving a set of three similar high-resolution
structures for the Mn4Ca cluster (Figure 2). This study, led
by scientists at the Physical Biosciences Division of Lawrence Berkeley
National Laboratory, involves an international collaborative effort between
groups at the Technische Universitńt and the Freie Universitńt, Berlin, and the
Max-Planck-Institut fŘr Bioanorganische Chemie in MŘlheim. The methodology and
the set up for collecting single crystal XAS data from PS II was developed in
collaboration with the Structural Biology group at SSRL.
|  |
|
Figure 2.
(A) FTs of polarized Mn EXAFS spectra from single crystals of PS
II in the S1
state. The FTs are from EXAFS spectra with the X-ray e-field vector aligned
parallel to the crystal unit cell axes of PSII [a (red curve), b (blue curve),
and c (green curve)]. Each of the three FT peaks characteristic of Mn EXAFS
from PSII is dichroic. FT peak I is from Mn-ligand backscattering; FT peak II
is from three Mn-Mn distances at 2.7 to 2.8 ┼; and FT peak III is from one
Mn-Mn and two Mn-Ca distances at 3.3 and 3.4 ┼, respectively. All Fourier peaks
appear at an apparent distance R' that is shorter than the actual distance R by
~0.5 ┼ due to a phase shift. The dichroism of the metal-to-metal distances
reflects the geometry of the Mn4Ca cluster.
(B) Structural models for the
Mn4Ca cluster in PS II from polarized EXAFS.
Each model is compatible with the polarized Mn EXAFS spectra of single crystals
of PS II. The Mn4 motif common to the three
structures is shown in the middle (top). The models are shown in the
orientation in which they should be placed in the PS II membrane according to
the axis system shown in the middle (bottom). Among the symmetry-related
orientation of each model, the particular orientations shown above were chosen
on the basis of their compatibility with the overall electron density and the
positioning of the protein ligands in the 3.0 ┼ resolution X-ray crystal
structure.6 The Mn atoms are shown in red. The distance between
MnC and MnD
atoms is ~2.8 ┼ (indicated by blue oxo bonds), and the distance between the
MnA
and MnB atoms, as well as the
MnB and MnC
atoms, is ~2.7 ┼. The distance
between MnB and MnD is ~3.3 ┼. The Ca atom (green sphere) is ~3.4 ┼ from
two Mn atoms. The bridging motif to Ca is not well defined by our experiments;
therefore, dashed lines connect the Ca atom to the two Mn atoms at ~3.4 ┼.
|
Combining polarized EXAFS and the electron density obtained from X-ray
crystallography data, the cluster was placed within PS II taking into account
the overall trend of the electron density of the metal site and the putative
ligands (Figure 3). This process successfully eliminates the
symmetry-related orientations that arise from the ~cos2q dependence of the EXAFS signal and the non-crystallographic
C2 symmetry of the monomers in the PS II dimer. Thus, the best-fit
ligand environment was obtained for all the models. The structure of
|  |
|
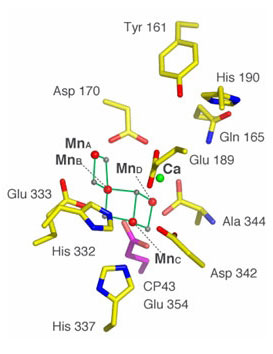
Figure 3. Placement of model II for the Mn4Ca cluster derived from
polarized Mn EXAFS in relation to the putative ligands obtained from the 3.0 ┼
resolution X-ray crystal structure.6 The spheres represent Mn (red), Ca
(green), and the bridging oxygen ligand atoms (gray). The assignment of ligands
is tentative because it is based on the electron density of the
Mn4Ca cluster,
and its immediate environment may be altered by X-ray damage.
| | | |
Mn4Ca cluster favoured in the present study contains structural features that
are unique and are likely to be important in mechanistically facilitating
water-oxidation. These models are unlike either the 3.0 or 3.5 ┼ resolution
X-ray structures, and other previously proposed models.
The current study demonstrates that the combination of XRD and polarized EXAFS
on single crystals has several advantages for unraveling structures of X-ray
damage-prone, redox-active metal sites in proteins. XRD structures at medium
resolution are sufficient to determine the overall shape and placement of the
metal site within the ligand sphere, and refinement using polarized EXAFS can
provide accurate metal-metal/ligand vectors. In addition, different
intermediate states of the active site (including different metal oxidation
states) can be studied, which may be difficult to study with XRD at high
resolution. The structural model from polarized EXAFS from the S1
state presented here, and from
the other S states, will provide a reliable foundation for the investigation of
the mechanism of photosynthetic water oxidation and for the design of
biomimetic catalysts for water splitting.
Primary Citation
Yano, J.; Kern, J.; Sauer, K.; Latimer, M. J.; Pushkar, Y.; Biesiadka, J.;
Loll, B.; Saenger, W.; Messinger, J.; Zouni, A.; Yachandra, V. K. Where Water
is Oxidized to Dioxygen: Structure of the Photosynthetic Mn4Ca
Cluster (2006) Science 314, 821-825.
References
-
Wydrzynski, T., Satoh, S. (Springer, Dordrecht, Netherlands, 2005).
-
Yachandra, V. K., Sauer, K. & Klein, M. P. (1996) Chem. Rev. 96,
2927-2950.
-
Zouni, A., Witt, H.-T., Kern, J., Fromme, P., Krau▀, N., Saenger, W. &
Orth, P. (2001) Nature 409, 739-743.
-
Kamiya, N. & Shen, J. R. (2003) Proc. Natl. Acad. Sci. USA
100, 98-103.
-
Ferreira, K. N., Iverson, T. M., Maghlaoui, K., Barber, J. & Iwata, S.
(2004) Science 303, 1831-1838.
-
Loll, B., Kern, J., Saenger, W., Zouni, A., Biesiadka, J. (2005) Nature
438, 1040.
-
Yano, J.; Kern, J.; Irrgang, K.-D.; Latimer, M. J.; Bergmann, U.;
Glatzel, P.; Pushkar, Y.; Biesiadka, J.; Loll, B.; Sauer, K.; Messinger, J.;
Zouni, A.; Yachandra, V. K. (2005) Proc. Natl. Acad. Sci. USA
102, 12047-12052.
|
| PDF
Version | | Lay Summary | |
Highlights Archive
|
| SSRL is supported
by the Department of Energy, Office of Basic Energy Sciences. The SSRL
Structural Molecular Biology Program is supported by the Department of Energy,
Office of Biological and Environmental Research, and by the National Institutes
of Health, National Center for Research Resources, Biomedical Technology
Program, and the National Institute of General Medical Sciences. |
|




