
The mammalian prion protein (PrP) folds into an alternative conformation
PrPSc to form the infectious entity responsible for human
Creutzfeldt-Jakob disease, bovine spongiform encephalopathy (mad cow disease),
scrapie in sheep, and several other mammalian CNS
disorders1. The mis-folded protein is the sole component of the
infectious prion. Prions can form amyloids, characterized by the formation of
long unbranched protein filaments, distinct staining properties, and a
structure of beta-strands approximately at right-angles
to the filament axis. This cross-beta structure is
indicated by meridional intensity at about 4.75 Å resolution in fiber
diffraction patterns. Although this characteristic diffraction feature has been
seen in many amyloids, until now it has not been observed for prions.
Amyloids have been implicated in more than forty diseases, including
neurodegenerative diseases such as Alzheimer's, Parkinson's, and
Creutzfeldt-Jakob diseases, as well as type II diabetes and other
non-neurological amyloidoses. There is increasing evidence that the propagation
of amyloid protein mis-folding, essentially a process of infection without any
requirement for a nucleic acid in the infecting material, is in principle the
same in functionally non-infectious diseases like Alzheimer's as it is in
overtly infectious prion diseases such as scrapie and mad cow disease.
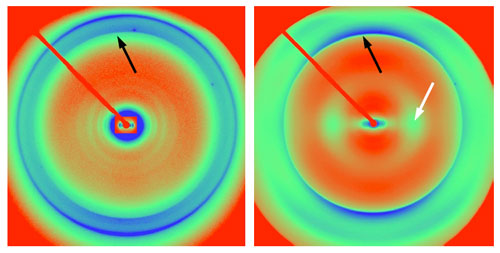
Figure 1. X-ray diffraction patterns from (left) brain-derived Syrian
hamster PrP 27-30 and (right) recombinant mouse PrP (89-230). The recombinant
hamster diffraction pattern is very similar to the mouse pattern. Black arrows
indicate cross-beta meridional diffraction at close to
4.8 Å resolution; white arrow indicates broad equatorial diffraction at
about 10.5 Å resolution, seen in recombinant diffraction patterns, but
not in patterns from brain-derived prions.
Diffraction data (Fig. 1) were obtained from fibers of hamster and mouse
brain-derived PrP 27-30, a proteolysed form of PrPSc that retains
full infectivity and about 65% of the prion protein. Diffraction from many
amyloids, and particularly from prions, is extremely weak because of both the
amyloid structure and the high degree of disorder often found in biological
amyloids. Interpretable diffraction required elaborate purification protocols
from hamster and mouse brains, carefully controlled conditions for fiber
formation2, and the exceptionally clean and intense
beam from SSRL Biological Small-Angle X-ray Scattering Beamline 4-2. Additional
data were obtained from the BioCARS beamline at the APS at Argonne National
Laboratory. The equatorial diffraction patterns from brain-derived prions were
characteristic of cylindrical structures, consistent with a beta-helical
structure such as has been proposed from electron microscopic and protein
folding considerations3. Weak meridional
diffraction in some patterns indicated an axial repeat of 19.2 Å, the
repeat expected from a four-stranded beta-sheet, again
supporting the proposed beta-helical structure.
Diffraction data were also obtained from fibers of recombinant mouse and
hamster PrP amyloid (Fig. 1). Although the recombinant amyloids have been found
to be infectious4, a remarkable achievement in
itself, the infectivity is much less than that of natural brain-derived prions.
The recombinant diffraction patterns were markedly different from those of
brain-derived prions. They were characterized by strong equatorial intensity at
approximately 10.5 Å, absent from brain-derived prions, and indicating
the presence of stacked beta-sheets. Diffraction
patterns calculated from the beta-helical structure and
a model stacked beta-sheet
structure5
strongly resembled the observed diffraction patterns for the brain-derived
prions and recombinant PrP amyloid respectively (Fig. 2).
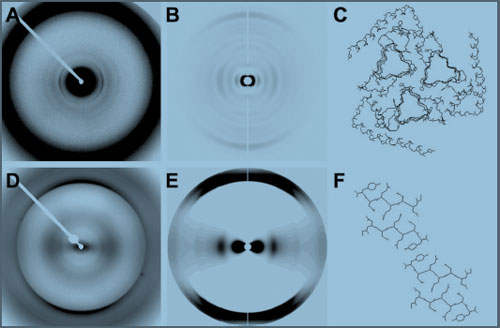
Figure 2. Observed and calculated diffraction patterns for beta-helical
and stacked-sheet models. (A) Experimental
diffraction pattern from hamster PrP 27-30. (B) Calculated diffraction
from a beta-helical model. (C) Model used to
calculate data in B. (D) Experimental diffraction pattern from
recombinant PrP amyloid. (E) Calculated diffraction from a stacked-sheet
model. (F) Model used to calculate data.
Diffraction from synthetic prions recovered from transgenic mice inoculated
with the recombinant PrP amyloid strongly resembled diffraction from naturally
occurring prions. A number of hypotheses might explain these observations. It
may be that only a small fraction of recombinant PrP amyloid has a
replication-competent conformation. Alternatively, the recombinant amyloid may
have to undergo a conformational maturation to acquire replication competency,
or inhibitory forms of recombinant amyloid may interfere with replication
during the initial transmission.
Primary Citation
Holger Wille, Wen Bian, Michele McDonald, Amy Kendall, David W. Colby, Lillian
Bloch, Julian Ollesch, Alexander L. Borovinskiy, Fred E. Cohen, Stanley B.
Prusiner, and Gerald Stubbs (2009) Natural and synthetic prion structure from
X-ray fiber diffraction. Proc. Natl. Acad. Sci. USA 106,
16990-95.
References
This work was supported in part by grants from the National Institutes of
Health (NS064, AG010770, and AG02132), the Fairchild Foundation, and the G.
Harold and Leila Y. Mathers Foundation. DWC was supported in part by a Jane
Coffin Childs postdoctoral fellowship.
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.