

The emergence of drug-resistant microbes represents a major impediment in the
treatment of bacterial infections. Resistance to first-choice drugs has become
problematic for respiratory infections, AIDS, tuberculosis, malaria and
diarrheal diseases, which are top killers among infectious agents. When second-
and third-choice drugs succumb to similar resistance, treatment options become
dire. As such, a major scientific priority in health-related research and
medicine is to identify new antimicrobial targets and to develop novel drugs
that keep infectious diseases in check according to global demand.
The mode of gene regulation by riboswitches represents a new paradigm for the
field but poses key questions: What is the minimal size of an aptamer? Is
the mode of metabolite recognition and expression-signal sequestration
universal for a given riboswitch class? In the current study, Joseph
Wedekind's lab at the University of Rochester (NY) solved the crystal structure
of the smallest, naturally occurring riboswitch aptamer known as
preQ1 (Fig. 1), which was derived from a hot spring bacterium
Thermoanaerobacter tengcongensis, and is bound to the guanine-like
metabolite preQ0 (Q0). Wedekind's group utilized beam
line 7-1 at SSRL, which was upgraded recently to allow multi-wavelength
anomalous diffraction and MAD phasing experiments. Dr. Wedekind and his team
accessed 7-1 remotely from Rochester, and have the distinction of solving the
Primary Citation
Spitale R.C., Torelli A.T, Krucinska J., Bandarian V., Wedekind J.E. (2009) The
structural basis for recognition of the PreQ0 metabolite by an
unusually small riboswitch aptamer domain. J. Biol. Chem. 284,
11012-6.
Blount, K. F., and Breaker, R. R. (2006) Nat. Biotechnol.
24, 1558-1564.
Roth, A., Winkler, W. C., Regulski, E. E., Lee, B. W., Lim, J., Jona, I.,
Barrick, J. E., Ritwik, A., Kim, J. N., Welz, R., Iwata-Reuyl, D., and Breaker,
R. R. (2007) Nat. Struct. Mol. Biol. 14, 308-317.
Iwata-Reuyl, D. (2003) Bioorg. Chem. 31, 24-43.
Editorial Note (2009) Minimal Recognition Required. J. Biol. Chem.
284, e99911.
This work was support in part by grants from the NIH (GM63162) and the donors
of the Petroleum Research Fund to JEW. RCS was supported in part by an Elon
Huntington Hooker graduate fellowship.
Riboswitches are a novel class of molecules that regulate the production of as
many as 4% of bacterial genes. In the classical view of gene expression,
instructions stored in DNA are copied into mRNAs, which make their way to the
ribosome where the genetic information is decoded as a protein. Groundbreaking
work by the Breaker lab at Yale and Nudler lab at NYU demonstrated in 2002 that
mRNA serves as more than a passive carrier of genetic information in this
decoding process. On the contrary, each group showed that key regions of
certain mRNAs are capable of "sensing" specific cellular metabolites. This
process occurs when conserved regions within the mRNA adopt a three-dimensional
fold within a so called 'aptamer domain' that embraces the smaller metabolite
molecule with high affinity - often at the nM level. The significance of this
recognition is that the 'folded' state of the mRNA sequesters critical signals
needed for successful protein production by the processes of transcription or
translation. Genetic regulation is achieved because the metabolite is often
produced by a protein whose mRNA harbors the riboswitch sequence, thus
providing direct feedback. When there is too much metabolite, the riboswitch
clenches its metabolite partner to inactivate expression. As levels of the
metabolite drop, the riboswitch relaxes its grip, exposing signals that favor
mRNA expression. Understanding how to manipulate a given riboswitch's "on" and
"off" states offers the prospect of a novel antimicrobial target, especially
when the associated genes encode metabolic intermediates that influence the
pathogenicity or survival of the bacterium.
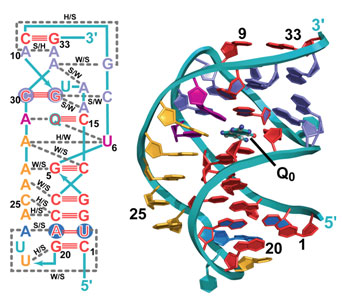
Figure 1:
Structure of the preQ1 riboswitch aptamer.
first MAD structure
on the improved line. The crystal structure revealed a highly compact RNA fold
in which more than 50% of all nucleotides are engaged in base triplet
interactions (Fig. 1). When the riboswitch twists into the well-known double
helix structure, the RNA bases align in the chain such that they stack on top
of each other, adding stability. Because Q0 is a modified form of
the guanine base, its presence in a solvent-sequestered riboswitch binding
pocket bridges a gap between otherwise unstacked RNA bases (Fig. 1), thus
contributing to the ~100 nM affinity of the aptamer domain for the metabolite.
Specificity for Q0 is achieved by "read out" of the metabolite by
C15 of the aptamer, which is reminiscent of the mode by which a G:C pair forms
in DNA (Fig. 2); other conserved bases assist in Q0 recognition,
such as U6 and A29. Perhaps most importantly, the new structure revealed that
the first base of the mRNA's ribosome binding site, G33, pairs with a C9 of the
riboswitch loop, which ties up the critical signal necessary for mRNA decoding.
Prior biochemical studies support the importance of maintaining the
preQ1
metabolic pathway for efficient bacterial growth, and evidence suggests that
the pathway is required for pathogenicity. Future investigations will focus on
whether the mode of metabolite recognition is universal or species specific,
which has implications for targeting various subgroups of bacteria and the
development of new antimicrobials.
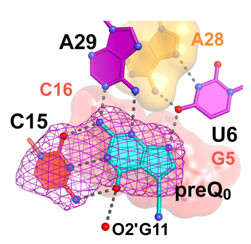
Figure 2: Read out of the preQ0 metabolite.
References
SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.