

Transcription is the first step and the key control point in the pathway of
gene expression. Transcriptional regulation underlies development, oncogenesis,
and other fundamental processes. The central enzyme in transcription is RNA
polymerase, in eukaryotic cells there are three forms of RNA polymerase,
designated I, II, and III (or A, B, and C), made up of 10-15 polypeptides. The
fundamental mechanism of transcription is conserved among cellular RNA
polymerases. Common features include an unwound region, or "transcription
bubble," of about 15 base pairs of the DNA template and some eight residues of
the RNA transcript hybridized with the DNA in the center of the bubble. These
enzymes are capable of both forward and retrograde movement ("backtracking") on
the DNA. Forward movement is favored by the binding of nucleoside triphosphates
(NTPs), while backtracking occurs especially when the enzyme encounters an
impediment, such as damaged DNA. All NTP's can bind the entry or "E" site,
whereas only an NTP matched for base pairing with the DNA template binds the A
(addition) site for addition to the growing RNA chain (1).
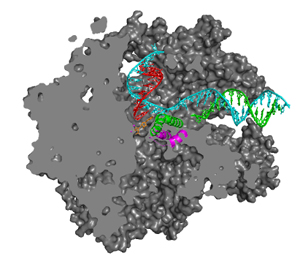
Figure 1. Cutaway view of the Pol II transcribing
complex. Template DNA, nontemplate DNA, RNA, GTP in the A site, are shown in
cyan, green, red, orange, respectively. The bridge helix (Rpb1 815- 848) is in
green; trigger loop (Rpb1 1065-1110) is in magenta and Mg2+ ions are
shown in magenta spheres. The pol II surface is shown in gray.
The way in which the correctly matched and positioned NTP is recognized and how
this recognition leads to catalysis remain obscure. The energies of base
pairing and stacking are insufficient for base selectivity, and the question
arises of why transient occupation of the A site by either incorrect NTP or
2'-dNTP substrates does not lead to erroneous RNA synthesis. Genetic and
biochemical studies have implicated two conserved polymerase domains, termed F
and G, in the transcription mechanism (2,3).
Structural studies have identified these two domains with elements adjacent to
the polymerase active site, termed the bridge helix (F) and trigger loop (G)
(4). In the x-ray structures of transcribing complexes,
however, no contact of these structural elements with NTP in the A or E sites
has been observed. The present work describes a series of pol II transcribing
complex structures that reveal such contacts and suggest the roles of these
domains in the transcription mechanism (Figure 1).
The trigger loop is a mobile element, allowing entry of NTP into the E and A
sites in conformations previously observed, and sealing off the A site in the
conformation reported here. Located beneath NTP in the A site, the trigger loop
directly contacts the base and b-phosphate, and
indirectly contacts 2'- and 3'-OH groups of the ribose sugar as well. Numerous
interactions with other pol II residues serve to configure and position the
trigger loop, so it reads out not only the chemical nature of the NTP but also
the parameters of the DNA-RNA hybrid helix in the A site. A well-defined
conformation of the trigger loop may be capable of readout to ┼ngstrom
precision. Inasmuch as the hybrid helix differs substantially from B-form DNA
(difference of 3 ┼ in minor groove width and 5.5 ┼ in root-mean- square
phosphorous positions), such readout would readily distinguish ribo from
deoxyribo NTPs, as well as providing powerful discrimination against
purine-purine and pyrimidine-pyrimidine mispairing.
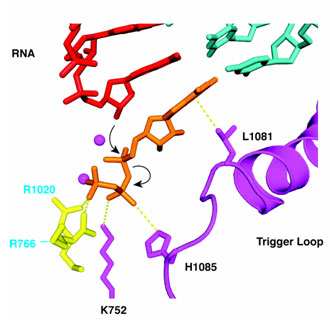
Figure 2. The pol II "trigger loop" forms a network
of interactions with a nucleoside triphosphate (NTP) in the active center. When
base, sugar, and phosphates are all correct, a histidine residue of the trigger
loop is aligned with the b-phosphate, facilitating
nucleophilic attack by the RNA 3'-OH and phosphodiester bond formation. In this
way, the trigger loop couples nucleotide selection to catalysis.
Two further features of trigger loop interaction may be crucial for
transcription. First, the contact of His1085 with the NTP b-phosphate noted
above may be key to catalysis. The distance between the imidazole N-H group and
b phosphate oxygen is about 3.5 ┼, optimal for hydrogen bonding or salt bridge
interaction. The protonated imidazole group would be expected to withdraw
electron density from the phosphate and facilitate SN2 attack of the
RNA 3'- terminal OH group, leading to phosphodiester bond formation (Figure
2). Second, trigger loop interaction with NTP in the A site is evidently poised
on the verge of stability, since the interaction could only be detected with
improved data quality and analysis. If any feature of the NTP or its location
is incorrect, the interaction will be lost.
The trigger loop may therefore couple nucleotide recognition to catalysis. In
the presence of matched rNTP in the A site, it will swing into position and
literally "trigger" phosphodiester bond formation (Figure 2). An incorrect NTP
in the A site will not support trigger loop interaction and so is unlikely to
undergo catalysis. When reaction with a correct NTP does occur, the release of
pyrophosphate disrupts contact with His1085, likely destabilizing trigger loop
interaction and freeing the DNA-RNA hybrid for translocation. Movement of the
trigger loop, coupled to that of the bridge helix (Figure 2), may contribute to
the translocation process (3,5).
Primary Citation
References
Wang, D., Bushnell, D.A., Westover, K.D., Kaplan, C.D., and Kornberg, R.D.
(2006) Structural basis of transcription: role of the trigger loop in substrate
specificity and catalysis. Cell 127, 941.
| SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. |