
 Raquel L. Lieberman,* Amy C. Rosenzweig,* and Timothy
L. Stemmler#
Raquel L. Lieberman,* Amy C. Rosenzweig,* and Timothy
L. Stemmler#
*Depts. of Biochemistry, Molecular Biology, and Cell Biology
and of Chemistry, Northwestern
University, Evanston, Illinois 60208, USA
#Dept. of Biochemistry and Molecular Biology, Wayne State
University, School of Medicine,
Detroit, Michigan 48201, USA.
Methane-oxidizing bacteria (methanotrophs) are extremely attractive from a chemist's perspective because these organisms convert methane to methanol. A detailed understanding of the conversion reaction, which occurs at ambient temperature and pressure, could lead to new methods for producing methanol, a potential alternative fuel source. Oxidation of methane is catalyzed by methane monooxygenase enzymes, which exist in both soluble (sMMO) and membrane-bound (pMMO) forms.1, 2 While the structure, biochemistry, and mechanism of sMMO are well understood,3 studies of pMMO are less advanced due to difficulties in solubilizing and purifying active protein. The Methylococcus capsulatus (Bath) pMMO comprises three polypeptides: a (~47 kDa), b (~24 kDa), and g (~22 kDa). It is not known how these three polypeptides are arranged in the pMMO holoenzyme.4, 5 The active site for methane oxidation is proposed to be located in the b subunit and might also involve the a subunit.6, 7
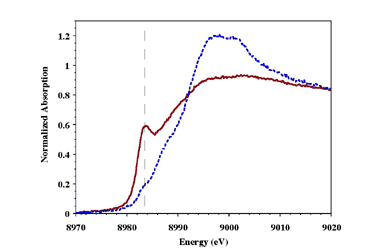 | |
| Figure 1: XANES Spectra of the as-isolated (dotted line) and reduced (solid line) pMMO. Dashed line identifies position of the 1s®4p transition. | |
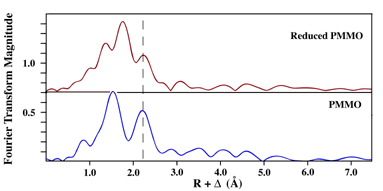 | |
| Figure 2: Fourier Transforms of as-isolated and reduced pMMO EXAFS Data. Dashed line identified metal-metal signal in the EXAFS. | |
We recently reported the structural characterization of copper centers in highly purified Methylococcus capsulatus pMMO using X-ray absorption spectroscopy.11 The as-isolated purified protein contains a mixture of Cu(I) and Cu(II), but can easily be reduced using dithionite (Figure 1). Fourier transforms of EXAFS data for purified and reduced purified pMMO indicate the presence of a rigid Cu-metal interaction at approximately 2.6 Å in both (Figure 2). Simulations of the EXAFS data for both indicate the presence of a distinct Cu-Cu interaction in pMMO at 2.57 Å, while the remaining nearest neighbor ligand environment around the copper is predominately constructed of oxygen and nitrogen based ligands.
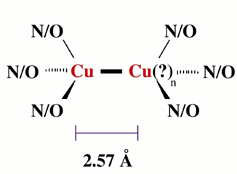 | |
| Figure 3: Proposed model for the Cu-metal interaction we observe in pMMO. The identity of the second metal ion has not yet been confirmed and could possibly be iron. The complex is not necessarily binuclear in metal (i.e., n=1 or 2, etc.) |
Based on our structural results, we propose the active site of pMMO is constructed partially (with 50% of the copper) of a multinuclear copper complex, although the nuclearity of the Cu cluster in pMMO is still unknown. Based on the short copper-copper distance, the active site in pMMO may resemble that seen in the catalytic CuZ cluster of nitrous oxide reductase.12 A similar complex has also been proposed by Lemos et al., in which a symmetric active site is constructed by two b subunits (Figure 3).13 Although additional studies are necessary to elucidate the structural details of the multinuclear center, these data provide the first evidence for a multinuclear cluster in pMMO.
References
- Murrell, J.C., McDonald, I.R. and Gilbert, B., (2000) Trends Microbiol., 8, 221-5.
- Stanley, S.H., Prior, S.D., Leak, D.J. and Dalton, H., (1983) Biotechnol. Lett., 6, 487-92.
- Merkx, M., D.A., K., Sazinsky, M.H., Blazyk, J.L., Muller, J. and Lippard, S.J., (2001) Agnew. Chem. Int. Ed., 40, 2782-807.
- Stolyar, S., Costello, A.M., Peeples, T.L. and Lidstrom, M.E., (1999) Microbiology, 145, 1235-44.
- Semrau, J.D., Chistoserdov, A., Lebron, J., Costello, A., Davagnino, J., Kenna, E., Holmes, A.J., Finch, R., Murrell, J.C., and Lidstrom, M.E., (1995) J. Bacteriol., 177, 3071-9.
- Cook, S.A. and Schiemke, A.K., (1996) J. Inorg. Biochem., 63, 273-84.
- Zahn, J.A. and DiSpirito, A.A., (1996) J. Bacteriol., 178, 1018-29.
- Basu, P., Katterle, B., Andersson, K.K. and Dalton, H., (2003) Biochem. J., 369, 412-27.
- Nguyen, H.H., Elliott, S.J., Yip, J.H. and Chan, S.I., (1998) J. Biol. Chem., 273, 7957-66.
- Nguyen, H.H.T., Nakagawa, K.H., Hedman, B., Elliott, S.J., Lidstrom, M.E., Hodgson, K.O., and Chan, S.I., (1996) J. Am. Chem. Soc., 118, 12766-76.
- Lieberman, R.L., Shrestha, D.B., Doan, P.E., Hoffman, B.M., Stemmler, T.L. and Rosenzweig, A.C., (2003) Proc. Natl. Acad. Sci. USA, 100, 3820-5.
- Brown, K., Djinovic-Carugo, K., Haltia, T., Cabrito, I., Saraste, M., Moura, J.J., Moura, I., Tegoni, M., and Cambillau, C., (2000) J. Biol. Chem., 275, 41133-6.
- Lemos, S.S., Yuan, H., Collins, M.L.P. and Antholine, W.E., (2002) Curr. Topics in Biophysics, 26, 43-8.
SSRL Highlights Archive