

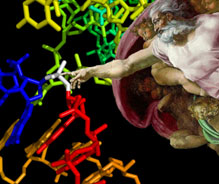
 | |
The L1 Ligase ribozyme at the moment of bond creation. |
The discovery 25 years ago that RNA can be enzymatic permits us to speculate that pre-biotic self-replicating molecules may have been RNAs (1,2). This is known as the "RNA World" hypothesis, and with the discovery of RNA catalysis, it is now possible to imagine a prebiotic 'RNA World' (or even one populated by early life forms) in which self-replicating ribozymes (RNA-based enzymes that possess the catalytic ability to copy themselves) accomplished both tasks, thus avoiding the potential "chicken-or-the-egg" conundrum (3).
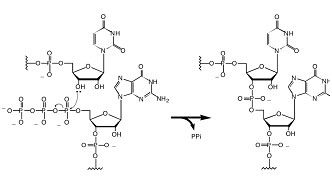 | |
The RNA nucleotide triphosphate ligation reaction required for RNA polymerization and self-replication. | |
In order to copy RNA, fragments or monomers that have 5'-triphosphates must be ligated together. This is true for modern polymerases, and is also the most likely mechanism by which a ribozyme self-replicase in an RNA World might function. Yet no one has found a modern natural ribozyme that catalyze this reaction (pictured right).
RNA in vitro evolution and selection has however enabled several
research groups to discover RNA sequences that can in fact catalyze the
required chemical reaction (shown above) for 5'-triphosphate RNA fragment
ligation, and one group has even produced a primitive but functional RNA-based
RNA polymerase ribozyme (4). This provides a Proof of
Principle that RNA is capable of such feats, and absent time travel, this is
likely the best we will be able to do.
The three-dimensional structure of the L1 ligase ribozyme. The ligation-site
phosphate is highlighted in white, and the catalytic magnesium ion in pink.
A close-up of the L1 ligase active site, revealing the positions of tightly
bound water (beige, W) and Mg2+ (pink), which in turn suggest a mechanism for
regiospecific 3' to 5' ligation.
In summary, the L1 ligase ribozyme (as well as other in vitro evolved
ligase ribozymes) is an empirical proof of principle that RNA is capable of
catalyzing the phosphodiester isomerization reaction that would be required of
any self-replicating RNA in a prebiotic RNA World. The three-dimensional
structure gives us our first glimpse of how this reaction is catalyzed by the
RNA in a regiospecific manner.
Primary Citation
References
As a first step to understanding what properties of ribozymes are of
fundamental importance to a plausible RNA World evolutionary scenerio, and how
RNA catalysts evolve in three-dimensional space, we have solved the structure
of an RNA Ligase Ribozyme that catalyzes the assembly of RNA from a
nucleotide-5'-triphosphate fragment.
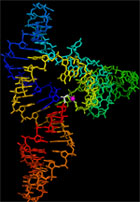
Dr. Michael Robertson in vitro selected a ligase ribozyme during his
Ph.D. with Dr. Andrew Ellington that regioselectively catalyzes a 5'-3' RNA
ligation reaction. He called this the L1 ligase. To better understand the
details of how this ribozyme folds into a structure that permits it to catalyze
this fundamental reaction, we have solved the structure of a catalytically
active variant of his original ribozyme, which we call L1X6c (left).
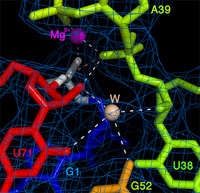
The three-dimensional structure reveals tertiary contacts, including an
invariant base triple, that help to position an essential Mg2+ at
the ligation site, along with a potentially catalytically-relevant
tightly-bound water molecule (right). Together with the RNA, these
moieties help to guide proper regiospecific assembly of a 3' to 5'
phosphodiester RNA backbone linkage. Specifically, the 2' and 3' protons of the
ribose substrate have similar pKas, and formation of an improper 2'
to 5' phosphodiester linkage would be equally probable to formation of a 3' to
5' linkage. The L1 ligase ribozyme appears to have evolved the capacity to
bind a water molecule within hydrogen bonding distance to the 2'-oxygen, which
might then lead to instant reprotonation of the aberrant nucleophile, should it
become deprotonated. This would enable the the 3' to 5' reaction to be
stereoselective in favor of the proper ligation product, i.e., the 3' to 5'
phosphodiester linkage observed in all natural nucleic acids.
Michael P. Robertson and William G. Scott. The structural basis for
ribozyme-catalyzed RNA assembly. Science, 2007, 315, 1549-1553.
| SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. |