
 Pierre Kennepohl1,2 and Edward Solomon1*
Pierre Kennepohl1,2 and Edward Solomon1*
1Department of Chemistry, Stanford University, Stanford, CA 94305
Electron transfer, or the act of moving an electron from one place to another, is amongst the simplest of chemical processes, yet certainly one of the most critical. The process of efficiently and controllably moving electrons around is one of the primary regulation mechanisms in biology. Without stringent control of electrons in living organisms, life could simply not exist. For example, photosynthesis and nitrogen fixation (to name but two of the most well-known biochemical activities) are driven by electron transfer processes. It is unsurprising, therefore, that much effort has been placed on understanding the fundamental principles that control and define the simple act of adding and/or removing electrons from chemical species.
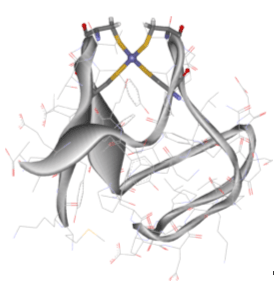 | |
| Figure 1. Three-dimensional structure of Desulfovibrio vulgaris Rubredoxin, a mononuclear iron-sulfur electron transport protein. The iron center is surrounded by four cysteine ligands that are attached by their sulfur atoms. |
Using small near-tetrahedral [Fe(SR)4]2-,1- models, we have probed the electronic structure of both the reduced (FeII) and oxidized (FeIII) complexes to obtain insights into their intrinsic redox properties and implications on the properties of Rubredoxins, a class of small electron transport proteins that contain a similar active site (Figure 1).5 An important question was whether electron transfer in these sites was well-described as a one-electron process. Generally, it is assumed that the removal and/or addition of a single electron is a rather simple process and that it does not significantly alter the electronic structure of a transition metal site. Another way of stating this is that we generally assume that electron transfer is a "one-electron" process - our approach to understanding electron transfer processes is rooted in this one-electron approximation. From studies on a related system, it seemed that this assumption might not hold.6, 7
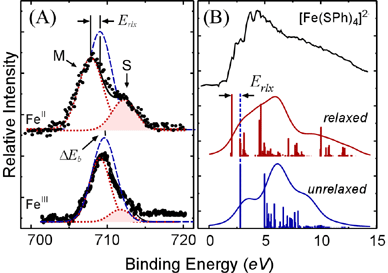 | |
| Figure 2. Core (A) and valence (B) PES data for the ferrous and ferric tetrathiolate model complexes. The data (black) are simulated using the one-electron model (blue) and including electronic relaxation (red). For the core data, the satellite feature (S) that results from electronic relaxation is shaded for emphasis. | |
This result is well-supported by theoretical results, which allow us to visualize the process and its influence. As we see from Figure 3, the overall electron transfer process looks very different than expected from the simple one-electron model. Using the one-electron model, the electron is removed mostly from the Fe atom, the ligands (the atoms attached to the metal) play only a very small role. However, the true situation is one that involves the ligands very heavily - electronic relaxation distributes the loss of the electron over a much wider volume. It is obvious at this point that relying on the one-electron approach to understand the electron transfer properties of these irons-sulfur sites is insufficient. For this reason, the importance of electronic relaxation on the intrinsic electron transfer properties of these iron-sulfur sites was explored, focusing on both the redox potentials and the rates constants for electron transfer.1
The experimental PES data from Figure 2 have already shown us that electronic relaxation is extremely important for the oxidation process.
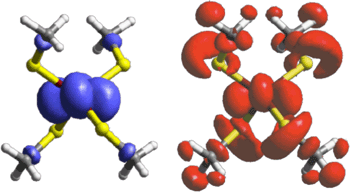 | |
| Figure 3. Theoretically-derived representation of the loss of an electron on going from [FeII(SCH3)4]2- to [FeIII(SCH3)4]1-. The top figure shows the calculated electron distribution using the one-electron formalism, whereas the bottom figure shows the electron distribution if electronic relaxation is allowed to take place. | |
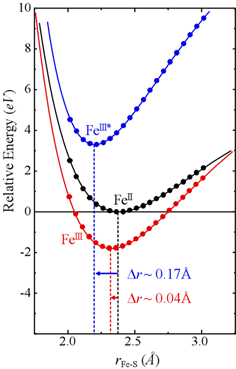 |
| Figure 4. Potential energy surfaces for the reduced (black) and oxidized (blue /red) forms of the iron site. The minimum in the plots indicate the average Fe-S bond distance in each case. The one-electron model (blue) predicts much larger bond distance changes than if we include electronic relaxation (red). |
A combination of synchrotron-based spectroscopy and theoretical methods has allowed us to demonstrate that the "one-electron" approach to understanding electron transfer can be misleading - a situation that is likely more general than this particular case. In so doing, we have obtained a new glimpse into the inherent redox properties of the mononuclear iron-sulfur active site in an important electron transfer protein, Rubredoxin. The simple fact that the electronic structure changes dramatically after removal of a single electron seems to be the determining factor that allows these proteins to do their job, and do it well.
| *Correspondence: | edward.solomon@stanford.edu |
| 2 Present address: | Department of Chemistry, University of British Columbia, Vancouver, BC, Canada V6T 1Z1 |
References:
- Kennepohl, P., and Solomon, E.I. (2003). Electronic Structure Contributions to Electron-Transfer Reactivity in Iron-Sulfur Active Sites: 1. Photoelectron Spectroscopic Determination of Electronic Relaxation. Inorganic Chemistry 42, 679-688.
- Kennepohl, P., and Solomon, E.I. (2003). Electronic Structure Contributions to Electron-Transfer Reactivity in Iron-Sulfur Active Sites: 2. Reduction Potentials. Inorganic Chemistry 42, 689-695.
- Kennepohl, P., and Solomon, E.I. (2003). Electronic Structure Contributions to Electron-Transfer Reactivity in Iron-Sulfur Active Sites: 3. Kinetics of Electron Transfer. Inorganic Chemistry 42, 696-708.
- Holm, R.H., Kennepohl, P., and Solomon, E.I. (1996). Structural and Functional Aspects of Metal Sites in Biology. Chemical Reviews 96, 2239-2314.
- Dauter, Z., Sieker, L.C., and Wilson, K.S. (1992). Refinement of rubredoxin from Desulfovibrio vulgaris at 1.0 A with and without restraints. Acta Crystallographica B 48, 42-59.
- Butcher, K.D., Didziulis, S.V., Briat, B., and Solomon, E.I. (1990). Variable-Photon-Energy Photoelectron Spectroscopic Studies of High-Spin d6 Tetrahedral FeCl42-: Electronic Relaxation Effects on Ionization. Inorganic Chemistry 29, 1626-1637.
- Butcher, K.D., Didziulis, S.V., Briat, B., and Solomon, E.I. (1990). Variable Photon Energy Photoelectron Spectroscopy on FeC14-: An Unusual Electronic Structure for High-Spin d5 Complexes. Journal of the American Chemical Society 112, 2231-2242.
This research was carried out at the Stanford Synchrotron Radiation Laboratory,
a national user facility operated by Stanford University on behalf of the U.S.
Department of Energy, Office of Basic Energy Sciences.
SSRL Highlights Archive