
by John Miao and Keith Hodgson
Protein
crystallography can routinely determine the 3D structure of protein molecules
at near atomic or atomic resolution. The bottleneck of this methodology
is to obtain sizable and good quality protein crystals. Overcoming the
crystallization difficulty requires the development of the new methodologies.
One approach is to use NMR to image protein molecules in solvent. However,
it is only applicable primarily to macromolecules in the lower molecular
weight range. Another approach under rapid development is single molecule
imaging using cryo electron microscopy (cryo-EM). The highest resolution
currently achievable by this technique is ~ 7 Å for highly symmetrical
viruses (1) and 11.5 Å for the asymmetrical ribosome
(2). The main limitations to achieving better resolution
by cryo-EM are radiation damage, specimen movement and low contrast. One
may ask why there is no such counterpart
for X-rays. One reason is the radiation damage problem. Due to the much
weaker interaction between matter with X-rays than with electrons, it requires
much higher radiation dose to achieve the same resolution by X-rays than
by electrons
(3). Another
is the difficulty of focussing X-rays. By using zone plates, the best
focus currently
achievable is ~ 30 nm for soft X-rays and ~ 100 nm
for hard X-rays (4).
|
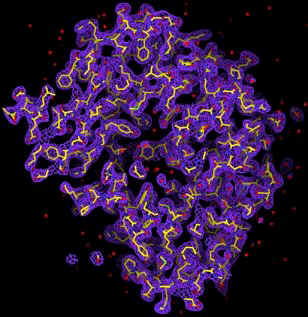
With the prospects of X-ray free electron lasers (X-FEL) (5, 6), the radiation damage problem could in principle be circumvented. Theoretical simulations by Hajdu and collaborators show that, within about 10 femtoseconds, biomolecules can withstand an X-ray intensity of ~ 3.8 x 106 photons/Å2 with minimal structural changes (7). A 2D diffraction pattern can hence be obtained from a single exposure of a biomolecule before the radiation manifests itself. From a set of such diffraction patterns covering 3D reciprocal space, the structure of the molecule can be obtained if phase information is available. The phase problem can be solved by using the oversampling method, which was first demonstrated by successfully converting an experimental diffraction pattern from a non-crystalline specimen (an array of sub-micron size dots) to the structure of the specimen (8). By combining the oversampling method with a simulated X-FEL, we, including David Sayre, have since carried out the computer modeling of imaging single biomolecules in 3-dimensions. We simulated an X-FEL with a wavelength of 1.5 Å, pulse flux of 2 x 1012 photons and a pulse length of 10 femtoseconds. The X-FEL was then focused down to a 100 nm diameter spot (9). We then processed a 3D diffraction pattern by utilizing the simulated X-FEL exposures and a large number of identical copies of single protein rubisco molecules. The 3D diffraction pattern extends to a resolution of 2.5 Å, which requires 106 identical copies of molecules. We then added Poisson noise to the diffraction pattern and also removed 3x3x3 pixel intensity at the center to simulate a beam stop. By employing the oversampling method, we can successfully reconstruct the 3D electron density map of the molecule directly from the noisy 3D diffraction pattern (10). The quality of the 3D electron map is comparable to that achieved from a conventional method. The promise of such experiments indeed offers one compelling motivation for the expedient construction of a next generation X-ray light source.
This work has been cited in several follow-up articles including:
- E. Lattman's commentary in PNAS (Proc. Natl. Acad. Sci. USA 98, 6535 (2001))
- The Bernstein Report on Biobusiness in BioCentury (June 11, 2001 p. A6)
- Nature Biotechnology (Nature Biotechnology 19, 739 (2001))
- Bottcher, B., Wynne, S. A. & Crowther, R. A. Nature 386, 88-91 (1997).
- Gabashvili, I. S., Agrawal, R. K., Spahn, C. M. T., Grassucci, R. A., Svergun, D. I., Frank, J & Penczek, P. Cell 100, 537-549 (2000).
- Henderson, R. Q. Rev. Biophys. 28, 171-193 (1995).
- Merner-Isle, W., Warwick, T. & Attwood, D. (eds) X-ray Microscopy (American Institute of Physics, 2000).
- Winick, H. J. Elec. Spec. Rel. Phenom. 75, 1-8 (1995).
- Wiik, B. H. Nucl. Inst. Meth. Phys. Res. B 398, 1-8 (1997).
- Neutze, R., Wouts, R., Spoel, D., Weckert, E. & Hajdu, J. Nature 406, 752-757 (2000).
- Miao, J., Charalambous, P., Kirz, J. & Sayre, D. Nature 400, 342-344 (1999).
- Tatchyn, R. LCLS Optics: Technological Issues and Scientific Opportunities in Proceedings of the Workshop on Scientific Applications of Short Wavelength Coherent Light Sources, SLAC, Stanford (1993).
- Miao, J., Hodgson, K. O. & Sayre, D. An Approach to 3-D Structures of Biomolecules by Using Single Molecule Diffraction Images, PNAS 98, 6641 (2001).
SSRL Highlights
Archive![]()
![]()
