 ATP-binding Cassette (ABC) transporters represent a large family of integral
membrane proteins, which are found in all organisms from mammals to bacteria.
These proteins transport substrates across a biological membrane powered by the
energy of adenosine triphosphate (ATP) hydrolysis. ABC transporters primarily
consist of two transmembrane domains (TMDs) and two nucleotide binding domains
(NBDs) located in the cytoplasm. While diverse with respect to physiological
function and TMD architecture, the highly conserved NBDs contain critical
sequence motifs for ATP binding and hydrolysis that suggest a common mechanism
by which these transporters translocate the substrate across the membrane
(1).
ATP-binding Cassette (ABC) transporters represent a large family of integral
membrane proteins, which are found in all organisms from mammals to bacteria.
These proteins transport substrates across a biological membrane powered by the
energy of adenosine triphosphate (ATP) hydrolysis. ABC transporters primarily
consist of two transmembrane domains (TMDs) and two nucleotide binding domains
(NBDs) located in the cytoplasm. While diverse with respect to physiological
function and TMD architecture, the highly conserved NBDs contain critical
sequence motifs for ATP binding and hydrolysis that suggest a common mechanism
by which these transporters translocate the substrate across the membrane
(1).
|  |
|
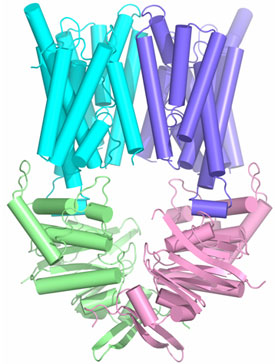
Figure 1. Structure of ABC transporter HI1470/1: HI1471, the membrane
spanning subunits are in cyan and purple and the nucleotide binding HI1470
subunits are in green and pink. | |
The HI1470/1 transporter from Haemophilus influenzae belongs to the
family of binding protein dependent bacterial ABC transporters that mediate the
uptake of metal-chelate species including heme and vitamin B12.
Using x-ray diffraction data collected at SSRL beam line 9-2, we have
determined the x-ray crystal structure of the intact, nucleotide-free HI1470/1
transporter by isomorphous and multi-wavelength anomalous diffraction methods
at 2.4 Å resolution (Figure 1). Although the overall architecture of the
intact HI1470/1 transporter resembles that of BtuCD (vitamin B12
uptake transporter), a comparison of the x-ray structures reveal differences in
tertiary and quaternary arrangements that may be functionally relevant
(2).
Each of the membrane spanning subunits HI1471 and BtuC contains a total of ten
transmembrane helices that are organized into two sets. The TMDs maintain a
tapered pathway across the membrane spanning region. Relative to a
structurally conserved core composed of seven of these helices, a twist of
~9°
about an axis perpendicular to the translocation pathway is required to
interconvert the TMDs of the two transporters. Together with the changes in
the three remaining helices, these conformational rearrangements result in a
translocation pathway that is closed to the periplasm and open to the cytoplasm
in HI1470/1, while the converse is observed in BtuCD. The pathways open to
opposite sides of the membrane such that HI1470/1 and BtuCD adopt inward and
outward facing conformations, respectively (Figure 2).
|  | |
Figure 2. Visualization of the permeation pathways of HI1470/1 and BtuCD
with the program HOLE (3). (A) The permeation pathway generated by the HI1471
TMD subunits (cyan and purple) is narrow at the periplasmic surface and open to
the cytoplasm. (B) In contrast, the pathway for BtuC (pink and red) is closed
at the cytoplasm and open to the periplasm.
| |
The differences observed between the structures of the HI1470/1 and BtuCD ABC
transporters involve relatively modest rearrangements and may serve as
structural models for inward and outward facing conformations relevant to the
alternating access mechanism of substrate translocation. The roles of binding
protein, ligand and particularly nucleotide binding and hydrolysis in driving
these conformational transitions remain crucial mechanistic issues that we are
presently exploring.
Primary Citation
H. W. Pinkett, A. T. Lee, P. Lum, K. P. Locher, D. C. Rees. (2007) An
Inward-Facing Conformation of a Putative Metal-Chelate-Type ABC Transporter.
Science 315, 373 - 377.
References
-
I. B. Holland, S. P. C. Cole, K. Kuchler, C. F. Higgins, ABC proteins: From
Bacteria to Man (Academic, London, 2003).
-
K. P. Locher, A. T. Lee, D. C. Rees, Science 296, 1091 (2002).
-
O. S. Smart, J. G. Neduvelil, X. Wang, B. A. Wallace, M. S. Sansom, J. Mol.
Graph. 14, 354 (1996).
|
| PDF
Version | | Lay Summary | |
Highlights Archive
|
| SSRL is supported
by the Department of Energy, Office of Basic Energy Sciences. The SSRL
Structural Molecular Biology Program is supported by the Department of Energy,
Office of Biological and Environmental Research, and by the National Institutes
of Health, National Center for Research Resources, Biomedical Technology
Program, and the National Institute of General Medical Sciences. |
|

 ATP-binding Cassette (ABC) transporters represent a large family of integral
membrane proteins, which are found in all organisms from mammals to bacteria.
These proteins transport substrates across a biological membrane powered by the
energy of adenosine triphosphate (ATP) hydrolysis. ABC transporters primarily
consist of two transmembrane domains (TMDs) and two nucleotide binding domains
(NBDs) located in the cytoplasm. While diverse with respect to physiological
function and TMD architecture, the highly conserved NBDs contain critical
sequence motifs for ATP binding and hydrolysis that suggest a common mechanism
by which these transporters translocate the substrate across the membrane
(1).
ATP-binding Cassette (ABC) transporters represent a large family of integral
membrane proteins, which are found in all organisms from mammals to bacteria.
These proteins transport substrates across a biological membrane powered by the
energy of adenosine triphosphate (ATP) hydrolysis. ABC transporters primarily
consist of two transmembrane domains (TMDs) and two nucleotide binding domains
(NBDs) located in the cytoplasm. While diverse with respect to physiological
function and TMD architecture, the highly conserved NBDs contain critical
sequence motifs for ATP binding and hydrolysis that suggest a common mechanism
by which these transporters translocate the substrate across the membrane
(1).

