
|
 |
|
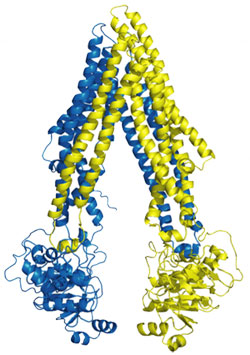
Figure 1. Structure of P-gp.
|
|
Many forms of cancer fail to respond to chemotherapy by acquiring multidrug
resistance (MDR), to which has been attributed the failure of treatment in over
90% of patients with metastatic cancer. Although MDR can have several causes,
one major form of resistance to chemotherapy has been correlated with the
presence of MDR "pumps" that actively transport drugs out of the cell. The most
prevalent of these MDR transporters is P-glycoprotein (P-gp), a member of the
adenosine triphosphate (ATP)-binding cassette (ABC) superfamily. P-glycoprotein
(P-gp) detoxifies cells by exporting hundreds of chemically unrelated toxins
but has been implicated in multidrug resistance in the treatment of cancers.
Most P-gp substrates are hydrophobic and partition into the lipid bilayer.
Thus, P-gp has been likened to a molecular "hydrophobic vacuum cleaner",
extracting substrates from the membrane and expelling them to promote multidrug
resistance. Although the structures of bacterial ABC importers and exporters
have been established, P-gp has only been characterized at low resolution by
electron microscopy. Substrate promiscuity is a hallmark of P-gp activity, thus
a structural description of poly-specific drug-binding is important for the
rational design of anticancer drugs and MDR inhibitors.
|  |
|
Figure 2. Model of substrate transport by P-gp.
|
|
Using data collected at SSRL Beam Lines 9-2, 11-1, and at ALS and APS, a
research team led by Geoffrey Chang from Scripps Research Institute determined
the three dimensional structure of P-gp and the inhibitor bound forms. The
x-ray structure of apo P-gp at 3.8 angstroms reveals an internal cavity of
6000 angstroms cubed with a 30 angstrom separation of the two
nucleotide-binding domains (Fig 1). The P-gp structures with cyclic peptide
inhibitors demonstrate distinct drug-binding sites in the internal cavity
capable of stereo selectivity that is based on hydrophobic and aromatic
interactions. Apo and drug-bound P-gp structures have portals open to the
cytoplasm and the inner leaflet of the lipid bilayer for drug entry. The
structure of P-gp represents a nucleotide-free inward-facing conformation
arranged as two "halves" with pseudo two-fold molecular symmetry spanning 136
Å perpendicular to and 70 Å in the plane of the bilayer.
The inward-facing structure does not allow substrate access from the outer
membrane leaflet nor the extracellular space and this conformation could
represent the molecule in a pre-transport state, because it is competent to
bind drug. This conformation likely represents an active state of P-gp because
protein recovered from crystals had significant drug-stimulated ATPase
activity. During the catalytic cycle, binding of ATP, stimulated by substrate,
likely causes a dimerization in the NBDs, which produces large structural
changes resulting in an outward-facing conformation similar that is
nucleotide-bound. ATP hydrolysis likely disrupts NBD dimerization and resets
the system back to inward facing and reinitiates the transport cycle.
Primary Citation
Stephen G. Aller, Jodie Yu, Andrew Ward, Yue Weng, Srinivas Chittaboina, Rupeng
Zhuo, Patina M. Harrell, Yenphuong T. Trinh, Qinghai Zhang, Ina L. Urbatsch,
Geoffrey Chang (2009) Structure of P-Glycoprotein Reveals a Molecular Basis for
Poly-specific Drug Binding. Science, 323, 1718-1722.
References
-
D. B. Longley, P. G. Johnston, J. Pathol. 205, 275 (2005).
-
F. J. Sharom, Pharmacogenomics 9, 105 (2008).
-
K. Hollenstein, D. C. Frei, K. P. Locher, Nature 446, 213 (2007).
-
H. W. Pinkett, A. T. Lee, P. Lum, K. P. Locher, D. C. Rees, Science
315, 373 (2007).
-
R. J. P. Dawson, K. P. Locher, Nature 443, 180 (2006).
-
A. Ward, C. L. Reyes, J. Yu, C. B. Roth, G. Chang, Proc. Natl. Acad. Sci.
U.S.A. 104, 19005 (2007).
|
|
This work was supported by grants from the Army (W81XWH-05-1-0316), NIH
(GM61905, GM078914, GM073197), the Beckman Foundation, the Skaggs Chemical
Biology Foundation, Jasper L. and Jack Denton Wilson Foundation, the Southwest
Cancer and Treatment Center, and the Norton B. Gilula Fellowship.
| PDF version | | Lay
Summary | |
Highlights Archive
|
SSRL is supported
by the Department of Energy, Office of Basic Energy Sciences. The SSRL
Structural Molecular Biology Program is supported by the Department of Energy,
Office of Biological and Environmental Research, and by the National Institutes
of Health, National Center for Research Resources, Biomedical Technology
Program, and the National Institute of General Medical Sciences.
|
|



