

Malaria is one of the most devastating parasitic diseases worldwide, amounting
to 300 to 500 million cases and ~2 million deaths per year (1). Multiple
species of Plasmodium infect the human host, the most important ones being
P. falciparum and P. vivax. Increased occurrence of
multi-drug resistant Plasmodium strains reflects the need for new effective
antimalarials. Human
infection starts when Anopheline mosquitoes inject sporozoites into the skin
during a blood meal. The sporozoites gain access to the blood stream and invade
liver cells where they develop and multiply. Upon rupture of the infected liver
cell, merozoites are released and rapidly enter red blood cells, where they
undergo schizogony (cell multiplication) and propagate the blood cycle of the
infection that causes the malaria symptoms (2).
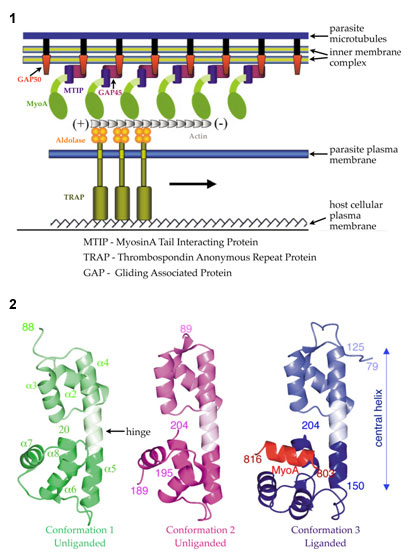
A multi-protein complex located in the narrow space between the parasite plasma
membrane and the microtubule-supported inner membrane complex empowers both
substrate-dependent gliding motility and host cell invasion in Plasmodium
(3).
This invasion machinery (Fig. 1) is highly conserved and required for leaving
and entering different types of host cells. Blocking one or more interactions
of the invasion machinery with small molecules inhibitors could provide
effective novel antimalarials. The current study provides atomic level insights
into a crucial protein interaction occurring in this essential multi-protein
assembly of the malaria parasite.
Using crystallographic data collected at SSRL beamline 9-2 and at the ALS, Wim
Hol's group have solved the structure of a complex of P. knowlesi
Myosin A-tail interacting protein (MTIP) and MyoA-tail to 2.6 Å
(P. knowlesi is a rodent
parasite. P. falciparum and P. knowlesi share 71% sequence
identity). The
crystals belonged to space group P63 and contained 3 different conformations of
MTIP in the asymmetric unit with one subunit bound with MyoA-tail (Fig. 2). The
Myosin A-tail interacting protein bridges between the membrane associated
proteins GAP45/GAP50 and the C-terminal tail of Myosin, which interacts with
short actin filaments (Fig. 1). The actin filaments are attached to the
glycolytic enzyme aldolase, which needs to be multimeric to connect actin and
TRAP. TRAP recognizes specific host cell receptors on red blood cells
initiating the process of invasion (Fig. 1).
A combination of hydrophilic and hydrophobic interactions is responsible for
forming the complex between the tail and the tail-binding protein (Fig. 2).
MyoA residues Gln-808 and His-810 make numerous hydrogen bonds with MTIP,
whereas electrostatic interactions mainly involve the first and third residues
from the conserved tribasic RKR motif spanning residues 812-814 in the
MyoA-tail. MyoA Arg-812 forms a salt bridge with MTIP Glu-180, whereas MyoA
Arg-814 is interacting with the side chain of MTIP Asp-202. Mutagenesis to Ala
of each of the two MyoA-tail Arg residues of the tribasic motif, and each of
the three hydrophobic MTIP-facing residues, results in a failure to interact
with MTIP, which is in complete agreement with the structure of the MyoA-MTIP
complex.
The viability as a drug target was tested by in vivo inhibition
experiments of P. falciparum cultures using the MyoA-tail. The binding
pocket for MyoA-tail provided by the C-terminal domain of MTIP displays
significant differences to the human homolog. These differences can be
exploited for structure based drug design of small molecules mimicking the
hydrophobic side chains of MyoA-tail yielding a higher binding affinity and
specificity.
This work was supported in part by NIH/NIGMS Grant 1P50 GM64655-01, Structural
Genomics of Pathogenic Protozoa and NIH Grant AI48226.
Primary Citation
References
Bosch, J., Turley, S., Daly, T. M., Bogh, S. M., Villasmil, M. L., Roach, C.,
Zhou, N., Morrisey, J. M., Vaidya, A. B., Bergman, L. W., et al. (2006).
Structure of the MTIP-MyoA complex, a key component of the malaria parasite
invasion motor. Proc. Natl. Acad. Sci. U. S. A. 103, 4852-4857
| SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. |