Vol. 19, No. 9 - May 2019
View the Archives

Science Highlights Winning the Fight against Influenza – Contact: Ian Wilson, The Scripps Research Institute Every year the flu vaccine contains a different formulation, due to multiple influenza virus strains and their ability to mutate. Scientists are working toward the universal flu vaccine, which would target conserved regions of the virus. Such a vaccine would be effective regardless of virus strain or genetic drift due to mutation, requiring no yearly updates. Current research focuses on the virus’s hemagglutinin (HA) stem region, which is targeted by our immune system’s broadly neutralizing antibodies (bnAbs). A team of researchers developed bnAbs to HA, which proved successful in preventing influenza infection in mice. Some of these are currently being evaluated for effectiveness in human clinical trials. The team is now focused on finding an effective small molecule based on their bnAbs, which would have the advantage of oral delivery. Read more...  Large-Scale Production of 119mTe and 119Sb for Radiopharmaceutical Applications – Contacts: Sharon Bone (SSRL) and Stosh Kozimor (LANL) Radioisotope therapies improve on traditional chemotherapies by being finely targeted to only the diseased cells and leaving surrounding healthy cells unharmed. A promising radioisotope for therapeutic uses is 119Sb, which releases low energy Auger electrons that can kill cancer cells. A problem with widespread use of drugs using 119Sb is its short half-life of around 38 hours. A team of scientists have developed a novel strategy for utilizing 119Sb. Read more... More SSRL ScienceIn a First, Researchers Identify Reddish Coloring in an Ancient Fossil – a 3-million-year-old Mouse Excerpt from May 21, 2019 SLAC News Article by Ali Sundermier
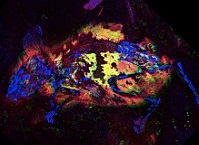
Researchers have for the first time detected chemical traces of red pigment in an ancient fossil – an exceptionally well-preserved mouse, not unlike today’s field mice, that roamed the fields of what is now the German village of Willershausen around 3 million years ago. The international collaboration, led by researchers at the University of Manchester in the U.K., used x-ray spectroscopy and multiple imaging techniques at SSRL and the Diamond Light Source to detect the delicate chemical signature of pigments in this long-extinct mouse. “Life on Earth has littered the fossil record with a wealth of information that has only recently been accessible to science,” says Phil Manning, a professor at Manchester who co-led the study. “A suite of new imaging techniques can now be deployed, which permit us to peer deep into the chemical history of a fossil organism and the processes that preserved its tissues. Where once we saw simply minerals, now we gently unpick the ‘biochemical ghosts’ of long extinct species.” The results were published today in Nature Communications. See SLAC News Release... Honors and AwardsStanford Faculty Elected to National Academy of Sciences Excerpt from April 30, 2019 Stanford News Article Stanford faculty Karla Kirkegaard, Todd Martinez and William Weis along with Mark Krasnow are now part of the National Academy of Sciences, an organization created in 1863 to advise the nation on issues related to science and technology.
Profs. Kirkegaard, Martinez and Weis are also part of the SSRL, LCLS or CryoEM user community at SLAC — in Weis' case, his research program spans across all three facilities. Weis studies molecular interactions that underlie the establishment and maintenance of cell and tissue structure using biochemical and biophysical methods. His lab’s specific areas of interest are the architecture and dynamics of intercellular adhesion junctions and signaling pathways that govern cell fate determination. The team also investigates carbohydrate-based cellular recognition and adhesion.
Kirkegaard’s work focuses on the impact of basic science discoveries on the transmission of viruses in infected hosts. She has combined her interests in biochemistry, cell biology and genetics in the study of RNA virology, using poliovirus and other positive-strand RNA viruses to understand the cell biology of viral infections and the genetics of viral variability.
Martinez studies physical and theoretical chemistry, bridging the gap between traditional molecular dynamics and quantum chemistry. He has developed interactive simulations to discover heretofore unknown chemical reactions with the goal of making molecular modeling both predictive and routine. Scholars are elected in recognition of their outstanding contributions to research. This year’s election brings the total of active academy members to 2,347. See Stanford News article Meeting SummaryAnother Successful RapiData Course for 2019! Arranging a macromolecular x-ray crystallography experiment into less than a week might seem impossible, but teaching the fundamentals and experimental tools certainly can be accomplished! That’s just what the Structural Molecular Biology (SMB) team at SSRL set out to do at SLAC in the form of RapiData 2019 - a workshop geared towards the collection and processing of x-ray data at synchrotrons.  For nearly two decades, the week-long RapiData course has aimed to educate and train young scientists in data collection and processing methods using state-of-the-art software and instrumentation. On May 5-10, 2019, co-organizers Clyde Smith, Silvia Russi, and Jeney Wierman of the SMB team brought together over a dozen specialized experts from around the world to give lectures and tutorials on the many aspects of a crystallography experiment to a class of 42 national and international students. Sessions on light sources and instrumentation, protein crystallography sample preparation, data collection and processing were also complemented with parallel techniques such as Cryo-EM and BioSAXS (as presented, respectively, by Wah Chiu and Thomas Weiss). In addition to the traditional instructional program, two prominent SSRL experimentalists gave pre-dinner guest lectures drawing direct connection between the experimental techniques being taught in the course and the impact of their results. Prof. Brian Kobilka of Stanford University concluded the first day of tutorials with a captivating presentation on structural approaches to understanding G protein coupled receptor (GPCR) signaling. Armed with humorous quips about the reality of data collection, Prof. Kobilka related his past GPCR experiments and framed the place of GPCR signaling in biology - a look into the work that awarded him a Nobel prize in Chemistry in 2012. As approximately half of all medications used today make use of GPCRs, his lectures underlined the importance of the tools the students were learning to perform through the RapiData course. To conclude the packed schedule of lectures and tutorials, Prof. James Fraser of U.C. San Francisco presented an intriguingly-titled talk, “Conformational Change We Can Believe In!" describing the influence of conformational change within protein structures at varying temperatures. His talk highlighted the renewed interest in room temperature data collection and gave emphasis to the new content from the added room temperature data collection lectures and tutorials within the RapiData circuit. As in previous years, IUCr and BioXFEL awarded travel scholarships to students on the final day. The organizers, instructors, tutors and students all shared their satisfaction and enthusiasm for the course, and look forward to continued RapiData courses at SLAC in the future. AnnouncementSubmit Structural Molecular Biology Multi-Technique Proposals by July 1, 2019
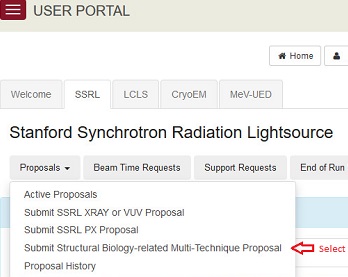
You can now submit a multi-technique proposal for structural biology-related projects that require the use of two or more scientific techniques available at SSRL (both x-ray and electron based). This pilot proposal mechanism is initially covering small angle x-ray scattering (SAXS), macromolecular crystallography (MC) and cryo-electron microscopy (CryoEM). In its permanent phase this program may expand to include additional techniques available at LCLS and spectroscopy and imaging methods at SSRL. Submit multi-technique proposals through the User Portal. Please provide a thorough justification for the requirement of using two or more techniques. The proposals will be reviewed by an ad hoc Proposal Review Panel comprised of members from the SSRL SMB and Cryo-EM PRPs. The deadline is July 1, 2019. To get started: Log into User Portal -> Select SSRL or CryoEM as the Facility -> Choose Submit Structural Biology-related Multi-Technique Proposal from SSRL Proposals or CryoEM Projects pulldown menu. More information about these techniques is available at SSRL's Structural Molecular Biology Program and the Stanford-SLAC Cryo-EM Facilities websites. EventsStanford-SLAC Cryo-EM Center (S2C2) Modeling Workshop - July 10-12, 2019 This workshop covers the basic principles and practical protocols to obtain atomic models based on cryo-EM density maps at near atomic resolution. See agenda Joint SSRL/LCLS Users’ Conference September 24-27, 2019 We look forward to welcoming you to this year's joint SSRL/LCLS Users' Meeting and workshops. This annual meeting is a unique opportunity to gather the light source community together in a single scientific event that includes numerous presentations in plenary, poster and parallel sessions. Participants are able to learn about current/future facility capabilities and the latest user research as well as to discuss science with colleagues from academia, research laboratories, and industry worldwide. We are planning several focused workshops including:
More details and registration information will be posted to the website soon. EMSL Integration 2019 – Plants, Soil and Aerosols: Interactions that tell stories of Ecosystems, Climate and National Security” October 8-10, 2019 announcement User Research AdministrationProposal Deadlines
See SSRL Proposal & Scheduling Guidelines Submit proposals through the User Portal. _____________________________________________________________________ The Stanford Synchrotron Radiation Lightsource (SSRL) is a third-generation light source producing extremely bright x-rays for basic and applied research. SSRL attracts and supports scientists from around the world who use its state-of-the-art capabilities to make discoveries that benefit society. SSRL, a U.S. DOE Office of Science national user facility, is a Directorate of SLAC National Accelerator Laboratory, operated by Stanford University for the U.S. Department of Energy Office of Science. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences. For more information about SSRL science, operations and schedules, visit http://www-ssrl.slac.stanford.edu. To unsubscribe from SSRL Headlines, just send an e-mail to listserv@slac.stanford.edu with "signoff ssrl-headlines" in the body. To subscribe, send an e-mail to listserv@slac.stanford.edu with "subscribe ssrl-headlines" in the body. Questions? Comments? Contact Lisa Dunn
|