The blueprint of life embedded in the genomic DNA, is first transcribed into mRNA, which is then translated into protein. In cells, the organelle for protein synthesis is the ribosome, which is comprised of two subunits. The mRNA is threaded through the small subunit of the ribosome and tRNA binds via base pairing with the triple-base codon in the mRNA, placing the attached amino acid in the large ribosomal subunit. The peptide linkage is then formed by joining amino acids at the transferase center of the large subunit. The two ribosomal subunits coordinate in reading through the mRNA sequence extending the corresponding polypeptide chain which then folds into a fully functional protein.
The accuracy of protein synthesis on the ribosome is rigorously maintained and the initiation of translation is carefully regulated. The initiation mechanism in eukaryotes is a complex process involving modification of mRNA (capping), initiator methionyl tRNA and up to a dozen initiation factors. The cell tightly controls the proteins being synthesized by adjusting the capping process as well as the availability of some of the initiation factors, ensuring normal growth under physiological conditions as well as responses to internal or external stresses.
Viruses do not have their own translation apparatus and have to use the host’s ribosome to synthesize their viral proteins. During viral infections, host cells down-regulate cap-dependent initiation as a defense, but some viruses utilize a structural feature in their RNA called an internal ribosome entry site (IRES) to bypass the requirement for a capped structure and for some of the initiation factors.
The mechanisms by which the IRESs operate differ among viruses and viral IRESs can be accordingly divided into several groups based on their secondary structural features and their functional needs for the subset of translation initiation factors. Considering the complexity of IRES initiation, theDicistroviradae intergenic region (IGR) IRES was chosen which uses the most streamlined mechanism known, directly binding to the ribosome without the need for an initiator tRNA or any initiation factors. Earlier structural and biochemical studies revealed a ribosome binding region as well as a tRNA mimic domain for this group of IRES RNAs. The crystal structure of two IGR IRES RNA domains was solved by the Kieft laboratory in 2006 and 2008 (1, 2). The Noller laboratory reported the first complete structure for the bacterial 70S ribosome bound with tRNA and mRNA at 5.5 Å resolution in 2001 and at higher resolution more recently (3, 4). Prompted by the universal phylogenetic conservation of the ribosome tRNA binding sites, an attempt was made to cocrystallize the IGR IRES tRNA mimic domain with the Thermus thermophilus 70S ribosome.
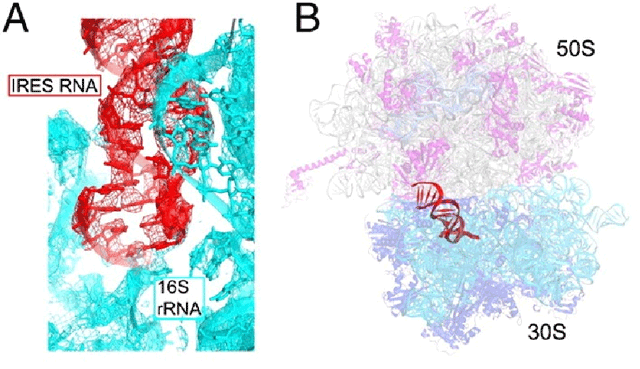
Based on their measured binding affinities of the IRES domains from 8 IGR viruseswith Thermus 70S ribosomes, Plautia stali intestine virus (PSIV) and Cricket paralysis virus (CrPV) were selected and successfully crystallized. More than 1000 crystals were prescreened at SSRL utilizing the Stanford Automated Mounting (SAM) system. Final x-ray diffraction datasets were collected for the 70S-CrPV complex at SSRL Beam Line 12-2 equipped with the PILATUS 6M detector, and at the Advanced Photon Source at Argonne National Laboratory for the 70S-PSIV complex. The structures were solved at 3.4 Å resolution for the 70S-CrPV and at 3.5Å for the 70S-PSIV tRNA mimic domains (Fig. 1A).
The two structures are very similar with the IRES domain sitting at the 30S P site (Fig. 1B). Comparing the two ribosome-bound IRES structures with the unbound CrPV crystal structure, overall they are very similar and include the tRNA aniticodon stem loop/mRNA codon mimic which reveals strong structural conservation across species and suggests this domain is pre-folded before ribosome binding.
In the P site of the small subunit of the ribosome-tRNA-mRNA complex structure, 7 contacts are observed: 3 between ribosomal RNA and mRNA and 4 between rRNA and tRNA. These 7 contacts are preserved in the IRES-ribosome structures and the 7 ribosome nucleotides involved are exactly the same as those in the tRNA complex (Fig. 2). For the 7 ribosomal nucleotides involved, 6 of them are identical in human 18S rRNA. This strongly suggests that the interactions observed in the P site of the IRES-70S ribosome complex may well correspond to reveal the actual structure of the complex formed between the IRES and the 80S ribosome.
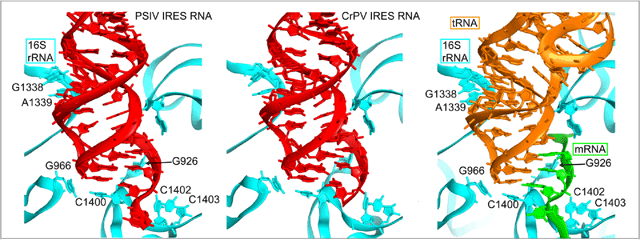
In the tRNA complex, a 25 degree kink projects the acceptor arm of the tRNA toward the 50S P site, placing its CCA end at the peptidyl transferase center. In contrast, extension of IRES domain stacks like a continuous RNA helix in these latest structures and is oriented toward the 50S subunit E site rather than the P site, avoiding any direct contact with the 50S subunit. This orientation of the IRES domain may mimic the P/E hybrid-state of the tRNA and favor A/P hybrid-state formation during translocation.
In summary, these structures combined with previous functional evidence show that the IGR IRES uses both direct structural mimicry and precise interactions with the ribosome to drive non-canonical translocation and initiation. In addition, this study shows how folded RNA structures can manipulate the ribosome, a strategy that might extend beyond viral IRESs to other noncoding RNAs.
This work was supported by Grants GM-17129 and GM-59140 from the NIH and MCB-723300 from the NSF (to H.F.N), Grants GM-72560 and GM-81346 from the NIH (to J.S.K.). J.S.K. is a Howard Hughes Medical Institute Early Career Scientist.
1. Jennifer Pfingsten, David Costantino and Jeffrey S. Kieft “Structural Basis for Ribosome Recruitment and Manipulation by a Viral IRES RNA” Science 314, 1450-1454, 2006
2. David Costantino, Jennifer Pfingsten, Robert Rambo and Jeffrey S Kieft “tRNA–mRNA mimicry drives translation initiation from a viral IRES” Nat Struct Mol Bio 15, 57-64, 2008
3. Marat M. Yusupov,Gulnara Zh. Yusupova, Albion Baucom, Kate Lieberman,Thomas N. Earnest, J. H. D. Cate and Harry F. Noller “Crystal Structure of the Ribosome at 5.5 Å Resolution” Science 292, 883-896, 2001
4. Martin Laurberg, Haruichi Asahara, Andrei Korostelev, Jianyu Zhu, Sergei Trakhanov and Harry F. Noller“Structural basis for translation termination on the 70S ribosome” Nature 454, 852-857, 2008
Zhu, J., Korostelev, A., Costantino, D. A., Donohue, J. P., Noller, H. F. and Kieft, J. S. (2011) Crystal structures of complexes containing domains from two viral IRES RNAs bound to the 70S ribosome Proc. Natl. Acad. Sci. USA, 108, 1839-1844.

